1B10
 
 | | SOLUTION NMR STRUCTURE OF RECOMBINANT SYRIAN HAMSTER PRION PROTEIN RPRP(90-231) , 25 STRUCTURES | | Descriptor: | PROTEIN (PRION PROTEIN) | | Authors: | James, T.L, Liu, H, Ulyanov, N.B, Farr-Jones, S. | | Deposit date: | 1998-11-25 | | Release date: | 1998-12-02 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a 142-residue recombinant prion protein corresponding to the infectious fragment of the scrapie isoform.
Proc.Natl.Acad.Sci.USA, 94, 1997
|
|
1C0Y
 
 | | SOLUTION STRUCTURE OF THE [AF]-C8-DG ADDUCT POSITIONED OPPOSITE DA AT A TEMPLATE-PRIMER JUNCTION | | Descriptor: | 2-AMINOFLUORENE, DNA (5'-D(*AP*AP*CP*GP*CP*TP*AP*CP*CP*AP*TP*CP*C)-3'), DNA (5'-D(*GP*GP*AP*TP*GP*GP*TP*AP*GP*C)-3') | | Authors: | Gu, Z, Gorin, A, Hingerty, B.E, Broyde, S, Patel, D.J. | | Deposit date: | 1999-07-19 | | Release date: | 1999-08-31 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structures of aminofluorene [AF]-stacked conformers of the syn [AF]-C8-dG adduct positioned opposite dC or dA at a template-primer junction.
Biochemistry, 38, 1999
|
|
1AB3
 
 | | RIBOSOMAL PROTEIN S15 FROM THERMUS THERMOPHILUS, NMR, 26 STRUCTURES | | Descriptor: | RIBOSOMAL RNA BINDING PROTEIN S15 | | Authors: | Berglund, H, Rak, A, Serganov, A, Garber, M, Hard, T. | | Deposit date: | 1997-02-03 | | Release date: | 1997-04-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the ribosomal RNA binding protein S15 from Thermus thermophilus.
Nat.Struct.Biol., 4, 1997
|
|
1B36
 
 | |
1ATW
 
 | | HAIRPIN WITH AGAU TETRALOOP, NMR, 3 STRUCTURES | | Descriptor: | RNA (5'-R(*GP*CP*UP*CP*CP*AP*GP*AP*UP*GP*GP*AP*GP*CP*G)-3') | | Authors: | Kang, H. | | Deposit date: | 1997-08-14 | | Release date: | 1998-02-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Primary Sequence at the Junction of Stem and Loop in RNA Hairpins Affects the Three-Dimensional Conformation in Solution
To be Published
|
|
1AU6
 
 | | SOLUTION STRUCTURE OF DNA D(CATGCATG) INTERSTRAND-CROSSLINKED BY BISPLATIN COMPOUND (1,1/T,T), NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | BIS(TRANS-PLATINUM ETHYLENEDIAMINE DIAMINE CHLORO)COMPLEX, DNA (5'-D(*CP*AP*TP*GP*CP*AP*TP*G)-3') | | Authors: | Yang, D, Van Boom, S.S.G.E, Reedijk, J, Van Boom, J.H, Farrell, N, Wang, A.H.-J. | | Deposit date: | 1997-09-11 | | Release date: | 1998-02-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A novel DNA structure induced by the anticancer bisplatinum compound crosslinked to a GpC site in DNA.
Nat.Struct.Biol., 2, 1995
|
|
1DFW
 
 | | CONFORMATIONAL MAPPING OF THE N-TERMINAL SEGMENT OF SURFACTANT PROTEIN B IN LIPID USING 13C-ENHANCED FOURIER TRANSFORM INFRARED SPECTROSCOPY (FTIR) | | Descriptor: | LUNG SURFACTANT PROTEIN B | | Authors: | Gordon, L.M, Lee, K.Y.C, Lipp, M.M, Zasadzinski, J.A, Walther, F.J, Sherman, M.A, Waring, A.J. | | Deposit date: | 1999-11-22 | | Release date: | 1999-12-10 | | Last modified: | 2024-02-07 | | Method: | INFRARED SPECTROSCOPY | | Cite: | Conformational mapping of the N-terminal segment of surfactant protein B in lipid using 13C-enhanced Fourier transform infrared spectroscopy.
J.Pept.Res., 55, 2000
|
|
1DY7
 
 | | Cytochrome cd1 Nitrite Reductase, CO complex | | Descriptor: | CARBON MONOXIDE, GLYCEROL, HEME C, ... | | Authors: | Sjogren, T, Svensson-Ek, M, Hajdu, J, Brzezinski, P. | | Deposit date: | 2000-01-28 | | Release date: | 2000-09-24 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Proton-Coupled Structural Changes Upon Binding of Carbon Monoxide to Cytochrome Cd(1): A Combined Flash Photolysis and X-Ray Crystallography Study
Biochemistry, 39, 2000
|
|
6BWZ
 
 | | SYSGYS from low-complexity domain of FUS, residues 37-42 | | Descriptor: | SYSGYS peptide from low-complexity domain of FUS | | Authors: | Hughes, M.P, Rodriguez, J.A, Sawaya, M.R, Cascio, D, Tamir, G, Eisenberg, D.S. | | Deposit date: | 2017-12-15 | | Release date: | 2018-04-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Atomic structures of low-complexity protein segments reveal kinked beta sheets that assemble networks.
Science, 359, 2018
|
|
5GHD
 
 | |
5GOX
 
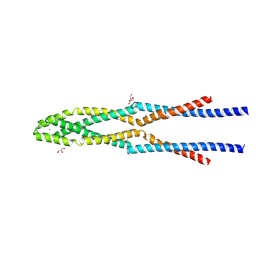 | | Eukaryotic Rad50 Functions as A Rod-shaped Dimer | | Descriptor: | DNA repair protein RAD50, GLYCEROL, ZINC ION | | Authors: | Park, Y.B, Hohl, M, Padjasek, M, Jeong, E, Jin, K.S, Krezel, A, Petrini, J.H.J, Cho, Y. | | Deposit date: | 2016-07-30 | | Release date: | 2017-02-01 | | Last modified: | 2017-03-15 | | Method: | X-RAY DIFFRACTION (2.405 Å) | | Cite: | Eukaryotic Rad50 functions as a rod-shaped dimer
Nat. Struct. Mol. Biol., 24, 2017
|
|
4IFI
 
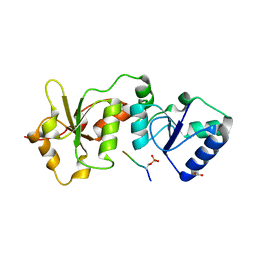 | |
4HFZ
 
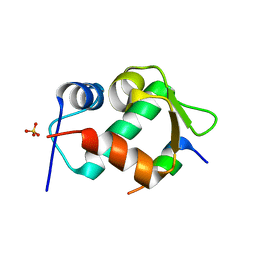 | | Crystal Structure of an MDM2/P53 Peptide Complex | | Descriptor: | Cellular tumor antigen p53, E3 ubiquitin-protein ligase Mdm2, SULFATE ION | | Authors: | Anil, B, Riedinger, C, Endicott, J.A, Noble, M.E.M. | | Deposit date: | 2012-10-05 | | Release date: | 2013-07-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.694 Å) | | Cite: | The structure of an MDM2-Nutlin-3a complex solved by the use of a validated MDM2 surface-entropy reduction mutant.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
8R63
 
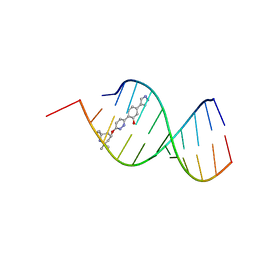 | | Solution structure of branaplam bound to the RNA duplex formed upon 5'-splice site recognition | | Descriptor: | 5-(1~{H}-pyrazol-4-yl)-2-[6-(2,2,6,6-tetramethylpiperidin-4-yl)oxypyridazin-3-yl]phenol, RNA (5'-R(*AP*UP*AP*CP*(PSU)P*(PSU)P*AP*CP*CP*UP*G)-3'), RNA (5'-R(P*GP*GP*AP*GP*UP*AP*AP*GP*UP*CP*U)-3') | | Authors: | Malard, F, Campagne, S. | | Deposit date: | 2023-11-20 | | Release date: | 2024-03-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The diversity of splicing modifiers acting on A-1 bulged 5'-splice sites reveals rules for rational drug design.
Nucleic Acids Res., 52, 2024
|
|
8R8P
 
 | |
8R1F
 
 | | Monomeric E6AP-E6-p53 ternary complex | | Descriptor: | Cellular tumor antigen p53, Ubiquitin-like protein SMT3,Protein E6, Ubiquitin-protein ligase E3A, ... | | Authors: | Sandate, C.R, Chakraborty, D, Kater, L, Kempf, G, Thoma, N.H. | | Deposit date: | 2023-11-01 | | Release date: | 2023-12-06 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.67 Å) | | Cite: | Structural insights into viral hijacking of p53 by E6 and E6AP
Biorxiv, 2023
|
|
8R1G
 
 | | Dimeric ternary structure of E6AP-E6-p53 | | Descriptor: | Cellular tumor antigen p53, Ubiquitin-like protein SMT3,Protein E6, Ubiquitin-protein ligase E3A, ... | | Authors: | Sandate, C.R, Chakrabory, D, Kater, L, Kempf, G, Thoma, N.H. | | Deposit date: | 2023-11-01 | | Release date: | 2023-12-06 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.99 Å) | | Cite: | Structural insights into viral hijacking of p53 by E6 and E6AP
Biorxiv, 2023
|
|
8R62
 
 | | Solution structure of Risdiplam bound to the RNA duplex formed upon 5'-splice site recognition | | Descriptor: | 7-(4,7-diazaspiro[2.5]octan-7-yl)-2-(2,8-dimethylimidazo[1,2-b]pyridazin-6-yl)-1~{H}-pyrido[1,2-a]pyrimidin-4-one, RNA (5'-R(*AP*UP*AP*CP*(PSU)P*(PSU)P*AP*CP*CP*UP*G)-3'), RNA (5'-R(P*GP*GP*AP*GP*UP*AP*AP*GP*UP*CP*U)-3') | | Authors: | Malard, F, Campagne, S. | | Deposit date: | 2023-11-20 | | Release date: | 2024-03-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The diversity of splicing modifiers acting on A-1 bulged 5'-splice sites reveals rules for rational drug design.
Nucleic Acids Res., 52, 2024
|
|
7VQQ
 
 | |
5LXD
 
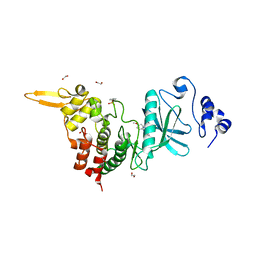 | | Crystal structure of DYRK2 in complex with EHT 1610 (compound 2) | | Descriptor: | 1,2-ETHANEDIOL, Dual specificity tyrosine-phosphorylation-regulated kinase 2, methyl 9-[(2-fluoranyl-4-methoxy-phenyl)amino]-[1,3]thiazolo[5,4-f]quinazoline-2-carboximidate | | Authors: | Chaikuad, A, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Besson, T, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-09-20 | | Release date: | 2016-10-26 | | Last modified: | 2017-01-11 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | An Unusual Binding Model of the Methyl 9-Anilinothiazolo[5,4-f] quinazoline-2-carbimidates (EHT 1610 and EHT 5372) Confers High Selectivity for Dual-Specificity Tyrosine Phosphorylation-Regulated Kinases.
J. Med. Chem., 59, 2016
|
|
5LXC
 
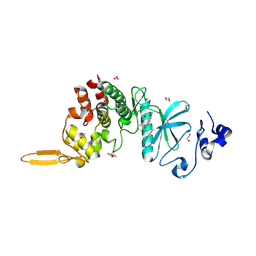 | | Crystal structure of DYRK2 in complex with EHT 5372 (Compound 1) | | Descriptor: | 1,2-ETHANEDIOL, Dual specificity tyrosine-phosphorylation-regulated kinase 2, methyl 9-[(2,4-dichlorophenyl)amino]-[1,3]thiazolo[5,4-f]quinazoline-2-carboximidate | | Authors: | Chaikuad, A, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Besson, T, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-09-20 | | Release date: | 2016-10-26 | | Last modified: | 2017-01-11 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | An Unusual Binding Model of the Methyl 9-Anilinothiazolo[5,4-f] quinazoline-2-carbimidates (EHT 1610 and EHT 5372) Confers High Selectivity for Dual-Specificity Tyrosine Phosphorylation-Regulated Kinases.
J. Med. Chem., 59, 2016
|
|
7XZZ
 
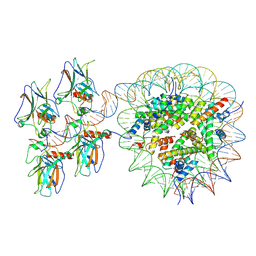 | | Cryo-EM structure of the nucleosome in complex with p53 | | Descriptor: | Cellular tumor antigen p53, DNA (169-MER), Histone H2A type 1-B/E, ... | | Authors: | Nishimura, M, Nozawa, K, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2022-06-03 | | Release date: | 2022-10-12 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (4.07 Å) | | Cite: | Structural basis for p53 binding to its nucleosomal target DNA sequence.
Pnas Nexus, 1, 2022
|
|
7XV4
 
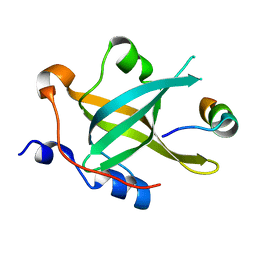 | | Crystal structure of RPA70N-ATRIP fusion | | Descriptor: | ATR-interacting protein, Replication protein A 70 kDa DNA-binding subunit | | Authors: | Wu, Y.Y, Zang, N, Fu, W.M, Zhou, C. | | Deposit date: | 2022-05-20 | | Release date: | 2023-06-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural characterization of human RPA70N association with DNA damage response proteins.
Elife, 12, 2023
|
|
7Z6H
 
 | | Structure of DNA-bound human RAD17-RFC clamp loader and 9-1-1 checkpoint clamp | | Descriptor: | Cell cycle checkpoint control protein RAD9A, Cell cycle checkpoint protein RAD1,Cell cycle checkpoint protein RAD17, Checkpoint protein HUS1, ... | | Authors: | Day, M, Oliver, A.W, Pearl, L.H. | | Deposit date: | 2022-03-11 | | Release date: | 2022-05-04 | | Last modified: | 2022-08-31 | | Method: | ELECTRON MICROSCOPY (3.59 Å) | | Cite: | Structure of the human RAD17-RFC clamp loader and 9-1-1 checkpoint clamp bound to a dsDNA-ssDNA junction.
Nucleic Acids Res., 50, 2022
|
|
7YXA
 
 | | XFEL crystal structure of the human sphingosine 1 phosphate receptor 5 in complex with ONO-5430608 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-[6-(2-naphthalen-1-ylethoxy)-2,3,4,5-tetrahydro-1H-3-benzazepin-3-ium-3-yl]butanoic acid, ... | | Authors: | Lyapina, E, Marin, E, Gusach, A, Orekhov, P, Gerasimov, A, Luginina, A, Vakhrameev, D, Ergasheva, M, Kovaleva, M, Khusainov, G, Khorn, P, Shevtsov, M, Kovalev, K, Okhrimenko, I, Bukhdruker, S, Popov, P, Hu, H, Weierstall, U, Liu, W, Cho, Y, Gushchin, I, Rogachev, A, Bourenkov, G, Park, S, Park, G, Huyn, H.J, Park, J, Gordeliy, V, Borshchevskiy, V, Mishin, A, Cherezov, V. | | Deposit date: | 2022-02-15 | | Release date: | 2022-08-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for receptor selectivity and inverse agonism in S1P 5 receptors.
Nat Commun, 13, 2022
|
|
