5MDN
 
 | | Structure of the family B DNA polymerase from the hyperthermophilic archaeon Pyrobaculum calidifontis | | Descriptor: | DNA polymerase, MAGNESIUM ION | | Authors: | Guo, J, Zhang, W, Coker, A.R, Wood, S.P, Cooper, J.B, Rashid, N, Akhtar, M. | | Deposit date: | 2016-11-12 | | Release date: | 2016-12-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the family B DNA polymerase from the hyperthermophilic archaeon Pyrobaculum calidifontis.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
3GYM
 
 | | Structure of Prostasin in Complex with Aprotinin | | Descriptor: | Pancreatic trypsin inhibitor, Prostasin | | Authors: | Spraggon, G, Hornsby, M, Shipway, A, Harris, J.L, Lesley, S.A. | | Deposit date: | 2009-04-03 | | Release date: | 2009-05-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Active site conformational changes of prostasin provide a new mechanism of protease regulation by divalent cations.
Protein Sci., 18, 2009
|
|
5LDV
 
 | | Crystal Structures of MOMP from Campylobacter jejuni | | Descriptor: | CALCIUM ION, MOMP porin, N-OCTANE, ... | | Authors: | Wallat, G.D, Ferrara, L.M.G, Moynie, L, Naismith, J.H. | | Deposit date: | 2016-06-28 | | Release date: | 2016-10-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | MOMP from Campylobacter jejuni Is a Trimer of 18-Stranded beta-Barrel Monomers with a Ca(2+) Ion Bound at the Constriction Zone.
J.Mol.Biol., 428, 2016
|
|
7E7F
 
 | | Human CYP11B1 mutant in complex with metyrapone | | Descriptor: | CHOLIC ACID, Cytochrome P450 11B1, mitochondrial, ... | | Authors: | Mukai, K, Sugimoto, H, Reiko, S, Matsuura, T, Hishiki, T, Kagawa, N. | | Deposit date: | 2021-02-26 | | Release date: | 2022-01-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Spatially restricted substrate-binding site of cortisol-synthesizing CYP11B1 limits multiple hydroxylations and hinders aldosterone synthesis.
Curr Res Struct Biol, 3, 2021
|
|
3RZW
 
 | |
6PAW
 
 | |
7WZS
 
 | |
2RP0
 
 | |
2RP1
 
 | |
6ZTM
 
 | | E. coli 70S-RNAP expressome complex in collided state without NusG | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Webster, M.W, Takacs, M, Weixlbaumer, A. | | Deposit date: | 2020-07-20 | | Release date: | 2020-09-16 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis of transcription-translation coupling and collision in bacteria.
Science, 369, 2020
|
|
7WR3
 
 | | Crystal structure of MBP-fused OspC3 in complex with calmodulin | | Descriptor: | Calmodulin-1, MBP-fused OspC3, NICOTINAMIDE, ... | | Authors: | Hou, Y.J, Zeng, H, Shao, F, Ding, J. | | Deposit date: | 2022-01-26 | | Release date: | 2023-01-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structural mechanisms of calmodulin activation of Shigella effector OspC3 to ADP-riboxanate caspase-4/11 and block pyroptosis.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7XN6
 
 | | Cryo-EM structure of CopC-CaM-caspase-3 with ADPR-deacylization | | Descriptor: | Arginine ADP-riboxanase CopC, Calmodulin-1, Caspase-3, ... | | Authors: | Zhang, K, Peng, T, Tao, X.Y, Tian, M, Li, Y.X, Wang, Z, Ma, S.F, Hu, S.F, Pan, X, Xue, J, Luo, J.W, Wu, Q.L, Fu, Y, Li, S. | | Deposit date: | 2022-04-28 | | Release date: | 2022-12-14 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Structural insights into caspase ADPR deacylization catalyzed by a bacterial effector and host calmodulin.
Mol.Cell, 82, 2022
|
|
8KA0
 
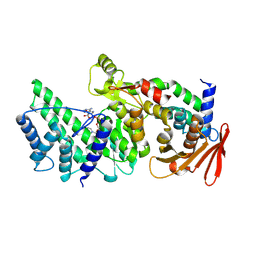 | | Crystal structure of Vibrio vulnificus RID-dependent transforming NADase domain (RDTND)/calmodulin-binding domain of Rho inactivation domain (RID-CBD) complexed with Ca2+-bound calmodulin and a nicotinamide adenine dinucleotide (NAD+) | | Descriptor: | CALCIUM ION, Calmodulin-2, GLYCEROL, ... | | Authors: | Lee, Y, Choi, S, Hwang, J, Kim, M.H. | | Deposit date: | 2023-08-02 | | Release date: | 2024-07-10 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Dissemination of pathogenic bacteria is reinforced by a MARTX toxin effector duet.
Nat Commun, 15, 2024
|
|
8K9Z
 
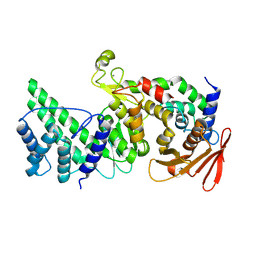 | | Crystal structure of Vibrio vulnificus RID-dependent transforming NADase domain (RDTND)/calmodulin-binding domain of Rho inactivation domain (RID-CBD) complexed with Ca2+-bound calmodulin | | Descriptor: | CALCIUM ION, Calmodulin-2, RDTND-RID CBD | | Authors: | Lee, Y, Choi, S, Hwang, J, Kim, M.H. | | Deposit date: | 2023-08-02 | | Release date: | 2024-07-10 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Dissemination of pathogenic bacteria is reinforced by a MARTX toxin effector duet.
Nat Commun, 15, 2024
|
|
8KA1
 
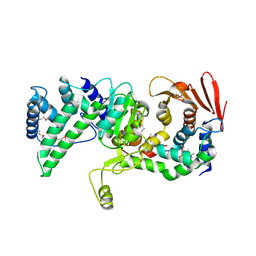 | | Crystal structure of Vibrio vulnificus RID-dependent transforming NADase domain (RDTND)/calmodulin-binding domain of Rho inactivation domain (RID-CBD) complexed with Ca2+-free calmodulin | | Descriptor: | Calmodulin-2, MAGNESIUM ION, RDTND-RID CBD | | Authors: | Lee, Y, Choi, S, Hwang, J, Kim, M.H. | | Deposit date: | 2023-08-02 | | Release date: | 2024-07-10 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Dissemination of pathogenic bacteria is reinforced by a MARTX toxin effector duet.
Nat Commun, 15, 2024
|
|
7VMB
 
 | |
6ZU1
 
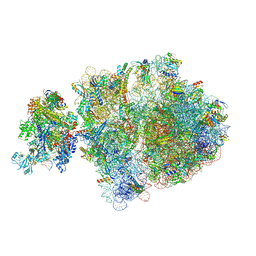 | | E. coli 70S-RNAP expressome complex in uncoupled state 2 | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Webster, M.W, Takacs, M, Weixlbaumer, A. | | Deposit date: | 2020-07-21 | | Release date: | 2020-09-16 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of transcription-translation coupling and collision in bacteria.
Science, 369, 2020
|
|
6ZTO
 
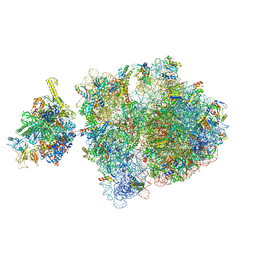 | | E. coli 70S-RNAP expressome complex in uncoupled state 1 | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Webster, M.W, Takacs, M, Weixlbaumer, A. | | Deposit date: | 2020-07-20 | | Release date: | 2020-09-23 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of transcription-translation coupling and collision in bacteria.
Science, 369, 2020
|
|
6ZTN
 
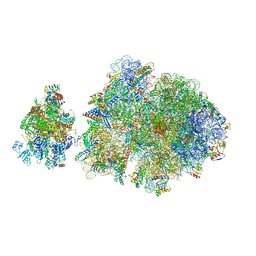 | | E. coli 70S-RNAP expressome complex in NusG-coupled state (42 nt intervening mRNA) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Webster, M.W, Takacs, M, Weixlbaumer, A. | | Deposit date: | 2020-07-20 | | Release date: | 2020-09-16 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural basis of transcription-translation coupling and collision in bacteria.
Science, 369, 2020
|
|
7XN4
 
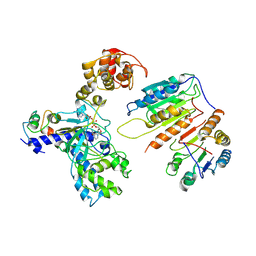 | | Cryo-EM structure of CopC-CaM-caspase-3 with NAD+ | | Descriptor: | Arginine ADP-riboxanase CopC, Calmodulin-1, Caspase-3, ... | | Authors: | Zhang, K, Peng, T, Tao, X.Y, Tian, M, Li, Y.X, Wang, Z, Ma, S.F, Hu, S.F, Pan, X, Xue, J, Luo, J.W, Wu, Q.L, Fu, Y, Li, S. | | Deposit date: | 2022-04-28 | | Release date: | 2022-12-14 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Structural insights into caspase ADPR deacylization catalyzed by a bacterial effector and host calmodulin.
Mol.Cell, 82, 2022
|
|
7XN5
 
 | | Cryo-EM structure of CopC-CaM-caspase-3 with ADPR | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, Arginine ADP-riboxanase CopC, Calmodulin-1, ... | | Authors: | Zhang, K, Peng, T, Tao, X.Y, Tian, M, Li, Y.X, Wang, Z, Ma, S.F, Hu, S.F, Pan, X, Xue, J, Luo, J.W, Wu, Q.L, Fu, Y, Li, S. | | Deposit date: | 2022-04-28 | | Release date: | 2022-12-14 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Structural insights into caspase ADPR deacylization catalyzed by a bacterial effector and host calmodulin.
Mol.Cell, 82, 2022
|
|
5WSU
 
 | | Crystal structure of Myosin VIIa IQ5-SAH in complex with apo-CaM | | Descriptor: | Calmodulin, Unconventional myosin-VIIa | | Authors: | Li, J, Chen, Y, Deng, Y, Lu, Q, Zhang, M. | | Deposit date: | 2016-12-08 | | Release date: | 2017-06-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Ca(2+)-Induced Rigidity Change of the Myosin VIIa IQ Motif-Single alpha Helix Lever Arm Extension
Structure, 25, 2017
|
|
5WSV
 
 | | Crystal structure of Myosin VIIa IQ5 in complex with Ca2+-CaM | | Descriptor: | CALCIUM ION, Calmodulin, SULFATE ION, ... | | Authors: | Li, J, Chen, Y, Deng, Y, Lu, Q, Zhang, M. | | Deposit date: | 2016-12-08 | | Release date: | 2017-06-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Ca(2+)-Induced Rigidity Change of the Myosin VIIa IQ Motif-Single alpha Helix Lever Arm Extension
Structure, 25, 2017
|
|
7WR4
 
 | | Crystal structure of OspC3-calmodulin-caspase-4 complex | | Descriptor: | Calmodulin-1, Caspase-4, OspC3 | | Authors: | Hou, Y.J, Zeng, H, Shao, F, Ding, J. | | Deposit date: | 2022-01-26 | | Release date: | 2023-01-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural mechanisms of calmodulin activation of Shigella effector OspC3 to ADP-riboxanate caspase-4/11 and block pyroptosis.
Nat.Struct.Mol.Biol., 30, 2023
|
|
5V7X
 
 | | Crystal Structure of Myosin 1b residues 1-728 with bound sulfate and Calmodulin | | Descriptor: | Calmodulin-1, SULFATE ION, Unconventional myosin-Ib | | Authors: | Zwolak, A, Shuman, H, Dominguez, R, Ostap, E.M. | | Deposit date: | 2017-03-20 | | Release date: | 2018-02-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.141 Å) | | Cite: | High-resolution cryo-EM structures of actin-bound myosin states reveal the mechanism of myosin force sensing.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
