1BYG
 
 | | KINASE DOMAIN OF HUMAN C-TERMINAL SRC KINASE (CSK) IN COMPLEX WITH INHIBITOR STAUROSPORINE | | Descriptor: | PROTEIN (C-TERMINAL SRC KINASE), STAUROSPORINE | | Authors: | Antson, A.A, Lamers, M.B.A.C, Scott, R.K, Williams, D.H, Hubbard, R.E. | | Deposit date: | 1998-10-14 | | Release date: | 1999-10-14 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the protein tyrosine kinase domain of C-terminal Src kinase (CSK) in complex with staurosporine.
J.Mol.Biol., 285, 1999
|
|
7DYV
 
 | | Human JMJD5 in complex with MN and 5-(benzylamino)pyridine-2,4-dicarboxylic acid. | | Descriptor: | 5-(benzylamino)pyridine-2,4-dicarboxylic acid, Bifunctional peptidase and arginyl-hydroxylase JMJD5, MANGANESE (II) ION | | Authors: | Nakashima, Y, Brewitz, L, Schofield, C.J. | | Deposit date: | 2021-01-23 | | Release date: | 2022-02-02 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | 5-Substituted Pyridine-2,4-dicarboxylate Derivatives Have Potential for Selective Inhibition of Human Jumonji-C Domain-Containing Protein 5.
J.Med.Chem., 66, 2023
|
|
7DYW
 
 | | Human JMJD5 in complex with MN and 5-((2-methoxybenzyl)amino)pyridine-2,4-dicarboxylic acid. | | Descriptor: | 5-((2-methoxybenzyl)amino)pyridine-2,4-dicarboxylic acid, Bifunctional peptidase and arginyl-hydroxylase JMJD5, MANGANESE (II) ION | | Authors: | Nakashima, Y, Brewitz, L, Schofield, C.J. | | Deposit date: | 2021-01-23 | | Release date: | 2022-02-02 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | 5-Substituted Pyridine-2,4-dicarboxylate Derivatives Have Potential for Selective Inhibition of Human Jumonji-C Domain-Containing Protein 5.
J.Med.Chem., 66, 2023
|
|
7DYT
 
 | | Human JMJD5 in complex with MN and 5-((4-methoxybenzyl)amino)pyridine-2,4-dicarboxylic acid. | | Descriptor: | 5-((4-methoxybenzyl)amino)pyridine-2,4-dicarboxylic acid, Bifunctional peptidase and arginyl-hydroxylase JMJD5, MANGANESE (II) ION | | Authors: | Nakashima, Y, Brewitz, L, Schofield, C.J. | | Deposit date: | 2021-01-23 | | Release date: | 2022-02-02 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | 5-Substituted Pyridine-2,4-dicarboxylate Derivatives Have Potential for Selective Inhibition of Human Jumonji-C Domain-Containing Protein 5.
J.Med.Chem., 66, 2023
|
|
7DYX
 
 | | Human JMJD5 in complex with MN and 5-((2-cyclopropylbenzyl)amino)pyridine-2,4-dicarboxylic acid. | | Descriptor: | 5-((2-cyclopropylbenzyl)amino)pyridine-2,4-dicarboxylic acid, Bifunctional peptidase and arginyl-hydroxylase JMJD5, MANGANESE (II) ION | | Authors: | Nakashima, Y, Brewitz, L, Schofield, C.J. | | Deposit date: | 2021-01-23 | | Release date: | 2022-02-02 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | 5-Substituted Pyridine-2,4-dicarboxylate Derivatives Have Potential for Selective Inhibition of Human Jumonji-C Domain-Containing Protein 5.
J.Med.Chem., 66, 2023
|
|
7DYU
 
 | | Human JMJD5 in complex with MN and 5-((4-phenylbutyl)amino)pyridine-2,4-dicarboxylic acid. | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5-(4-phenylbutylamino)pyridine-2,4-dicarboxylic acid, Bifunctional peptidase and arginyl-hydroxylase JMJD5, ... | | Authors: | Nakashima, Y, Brewitz, L, Schofield, C.J. | | Deposit date: | 2021-01-23 | | Release date: | 2022-02-02 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | 5-Substituted Pyridine-2,4-dicarboxylate Derivatives Have Potential for Selective Inhibition of Human Jumonji-C Domain-Containing Protein 5.
J.Med.Chem., 66, 2023
|
|
3UU9
 
 | | Structure of the free TvNiRb form of Thioalkalivibrio nitratireducens cytochrome c nitrite reductase | | Descriptor: | CALCIUM ION, Eight-heme nitrite reductase, HEME C, ... | | Authors: | Trofimov, A.A, Polyakov, K.M, Tikhonova, T.V, Tikhonov, A.V, Dorovatovskii, P.V, Popov, V.O. | | Deposit date: | 2011-11-28 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Covalent modifications of the catalytic tyrosine in octahaem cytochrome c nitrite reductase and their effect on the enzyme activity.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3GWK
 
 | | Structure of the homodimeric WXG-100 family protein from Streptococcus agalactiae | | Descriptor: | Putative uncharacterized protein SAG1039, SULFATE ION | | Authors: | Poulsen, C, Gries, F, Wilmanns, M, Song, Y.H. | | Deposit date: | 2009-04-01 | | Release date: | 2010-09-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | WXG100 protein superfamily consists of three subfamilies and exhibits an alpha-helical C-terminal conserved residue pattern.
Plos One, 9, 2014
|
|
4FZJ
 
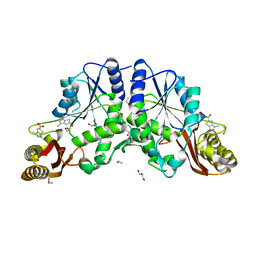 | | Pantothenate synthetase in complex with 1,3-DIMETHYL-1H-THIENO[2,3-C]PYRAZOLE-5-CARBOXYLIC ACID | | Descriptor: | 1,2-ETHANEDIOL, 1,3-dimethyl-1H-thieno[2,3-c]pyrazole-5-carboxylic acid, ETHANOL, ... | | Authors: | Silvestre, H.L, Blundell, T.L, Abell, C, Ciulli, A. | | Deposit date: | 2012-07-06 | | Release date: | 2013-06-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Integrated biophysical approach to fragment screening and validation for fragment-based lead discovery.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
5VGD
 
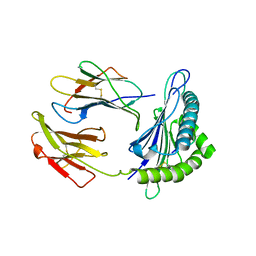 | | Crystal Structure of HLA-C*0501 in complex with SAE | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, Cw-5 alpha chain, ... | | Authors: | Gras, S, Rossjohn, J. | | Deposit date: | 2017-04-11 | | Release date: | 2017-05-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Structural and regulatory diversity shape HLA-C protein expression levels.
Nat Commun, 8, 2017
|
|
3VDM
 
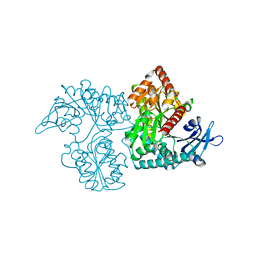 | | Crystal Structure of VldE, the pseudo-glycosyltransferase which catalyzes non-glycosidic C-N coupling in Validamycin A biosynthesis | | Descriptor: | VldE | | Authors: | Cavalier, M.C, Yim, Y.-S, Asamizu, S, Neau, D, Mahmud, T, Lee, Y.-H. | | Deposit date: | 2012-01-05 | | Release date: | 2013-01-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Crystal Structure of VldE, the pseudo-glycosyltransferase which catalyzes non-glycosidic C-N coupling in Validamycin A biosynthesis
To be Published
|
|
3TCX
 
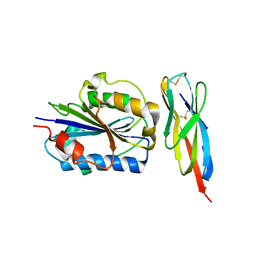 | | Structure of Engineered Single Domain ICAM-1 D1 with High-Affinity aL Integrin I Domain of Native C-Terminal Helix Conformation | | Descriptor: | Integrin alpha-L, Intercellular adhesion molecule 1, MAGNESIUM ION | | Authors: | Kang, S, Kim, C.U, Gu, X, Owens, R.M, van Rijn, S.J, Boonyaleepun, V, Mao, Y, Springer, T.A, Jin, M.M. | | Deposit date: | 2011-08-09 | | Release date: | 2011-08-31 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structure of Engineered Single Domain ICAM-1 D1 with High-Affinity L Integrin I Domain of Native C-Terminal Helix Conformation
To be Published
|
|
6TLG
 
 | | Ligand-free state of human 14-3-3 sigma isoform | | Descriptor: | 14-3-3 protein sigma, DI(HYDROXYETHYL)ETHER, SULFATE ION | | Authors: | Tassone, G, Pozzi, C, Mangani, S. | | Deposit date: | 2019-12-02 | | Release date: | 2020-03-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Identification of Phosphate-Containing Compounds as New Inhibitors of 14-3-3/c-Abl Protein-Protein Interaction.
Acs Chem.Biol., 15, 2020
|
|
5VOJ
 
 | |
6EI2
 
 | | Crystal Structure of HLA-A68 presenting a C-terminally extended peptide | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, CADMIUM ION, ... | | Authors: | Picaud, S, Guillaume, P, Pike, A.C.W, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Gfeller, D, Filippakopoulos, P. | | Deposit date: | 2017-09-16 | | Release date: | 2017-10-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Crystal Structure of HLA-A68 presenting a C-terminally extended peptide
To Be Published
|
|
1C6V
 
 | | SIV INTEGRASE (CATALYTIC DOMAIN + DNA BIDING DOMAIN COMPRISING RESIDUES 50-293) MUTANT WITH PHE 185 REPLACED BY HIS (F185H) | | Descriptor: | PROTEIN (SIU89134), PROTEIN (SIV INTEGRASE) | | Authors: | Chen, Z, Yan, Y, Munshi, S, Li, Y, Zruygay-Murphy, J, Xu, B, Witmer, M, Felock, P, Wolfe, A, Sardana, V, Emini, E.A, Hazuda, D, Kuo, L.C. | | Deposit date: | 1999-12-21 | | Release date: | 2000-12-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | X-ray structure of simian immunodeficiency virus integrase containing the core and C-terminal domain (residues 50-293)--an initial glance of the viral DNA binding platform.
J.Mol.Biol., 296, 2000
|
|
1SZM
 
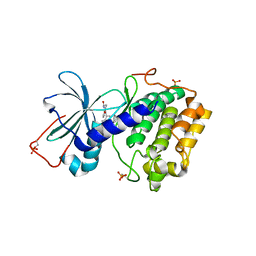 | | DUAL BINDING MODE OF BISINDOLYLMALEIMIDE 2 TO PROTEIN KINASE A (PKA) | | Descriptor: | 3-(1H-INDOL-3-YL)-4-{1-[2-(1-METHYLPYRROLIDIN-2-YL)ETHYL]-1H-INDOL-3-YL}-1H-PYRROLE-2,5-DIONE, cAMP-dependent protein kinase, alpha-catalytic subunit | | Authors: | Gassel, M, Breitenlechner, C.B, Koenig, N, Huber, R, Engh, R.A, Bossemeyer, D. | | Deposit date: | 2004-04-06 | | Release date: | 2004-06-01 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The protein kinase C inhibitor bisindolyl maleimide 2 binds with reversed orientations to different conformations of protein kinase a.
J.Biol.Chem., 279, 2004
|
|
3S0D
 
 | | Apis mellifera OBP 14 in complex with the citrus odorant citralva (3,7-dimethylocta-2,6-dienenitrile) | | Descriptor: | (2Z)-3,7-dimethylocta-2,6-dienenitrile, OBP14 | | Authors: | Spinelli, S, Lagarde, A, Iovinella, I, Tegoni, M, Pelosi, P, Cambillau, C. | | Deposit date: | 2011-05-13 | | Release date: | 2011-11-30 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Crystal structure of Apis mellifera OBP14, a C-minus odorant-binding protein, and its complexes with odorant molecules.
Insect Biochem.Mol.Biol., 42, 2012
|
|
3S0B
 
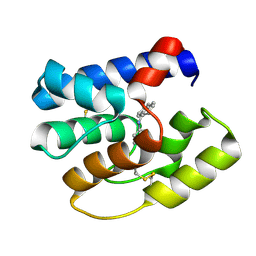 | | Apis mellifera OBP14 in complex with the fluorescent probe 1-N-phenylnaphthylamine (NPN) | | Descriptor: | N-phenylnaphthalen-1-amine, OBP14 | | Authors: | Spinelli, S, Lagarde, A, Iovinella, I, Tegoni, M, Pelosi, P, Cambillau, C. | | Deposit date: | 2011-05-13 | | Release date: | 2011-11-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Crystal structure of Apis mellifera OBP14, a C-minus odorant-binding protein, and its complexes with odorant molecules.
Insect Biochem.Mol.Biol., 42, 2012
|
|
7KHA
 
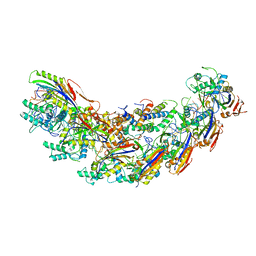 | | Cryo-EM Structure of the Desulfovibrio vulgaris Type I-C Apo Cascade | | Descriptor: | CRISPR-associated protein, CT1133 family, CT1134 family, ... | | Authors: | O'Brien, R, Wrapp, D, Bravo, J.P.K, Schwartz, E, Taylor, D. | | Deposit date: | 2020-10-20 | | Release date: | 2020-11-11 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Structural basis for assembly of non-canonical small subunits into type I-C Cascade.
Nat Commun, 11, 2020
|
|
2K8Z
 
 | | Dimeric solution structure of the DNA loop d(TCGTTGCT) | | Descriptor: | 5'-D(*TP*CP*GP*TP*TP*GP*CP*T)-3' | | Authors: | Viladoms, J, Escaja, N, Frieden, M, Gomez-Pinto, I, Pedroso, E, Gonzalez, C. | | Deposit date: | 2008-09-29 | | Release date: | 2009-04-28 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Self-association of short DNA loops through minor groove C:G:G:C tetrads.
Nucleic Acids Res., 37, 2009
|
|
5GMJ
 
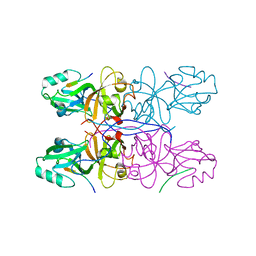 | | Crystal Structure of GRASP55 GRASP domain in complex with JAM-B C-terminus | | Descriptor: | Golgi reassembly-stacking protein 2, Junctional adhesion molecule B | | Authors: | Shi, N, Shi, X, Morelli, X, Betzi, S, Huang, X. | | Deposit date: | 2016-07-14 | | Release date: | 2017-05-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.986 Å) | | Cite: | Genetic, structural, and chemical insights into the dual function of GRASP55 in germ cell Golgi remodeling and JAM-C polarized localization during spermatogenesis
PLoS Genet., 13, 2017
|
|
5E6O
 
 | | Crystal structure of C. elegans LGG-2 bound to an AIM/LIR motif | | Descriptor: | Protein lgg-2, TRP-GLU-GLU-LEU | | Authors: | Qi, X, Ren, J.Q, Wu, F, Zhang, H, Feng, W. | | Deposit date: | 2015-10-10 | | Release date: | 2016-01-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis of the Differential Function of the Two C. elegans Atg8 Homologs, LGG-1 and LGG-2, in Autophagy
Mol.Cell, 60, 2015
|
|
4HZL
 
 | |
1V0C
 
 | | Structure of AAC(6')-Ib in complex with Kanamycin C and AcetylCoA. | | Descriptor: | AAC(6')-IB, ACETYL COENZYME *A, CALCIUM ION, ... | | Authors: | Vetting, M.W, Park, C.H, Hedge, S.S, Hooper, D.C, Blanchard, J.S. | | Deposit date: | 2008-03-20 | | Release date: | 2008-09-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mechanistic and Structural Analysis of Aminoglycoside N-Acetyltransferase Aac(6')-Ib and its Bifunctional, Fluoroquinolone-Active Aac(6')-Ib-Cr Variant.
Biochemistry, 47, 2008
|
|
