4N40
 
 | | Crystal structure of human Epithelial cell-transforming sequence 2 protein | | Descriptor: | Protein ECT2 | | Authors: | Zou, Y, Shao, Z.H, Li, F.D, Gong, D, Wang, C, Gong, Q, Shi, Y. | | Deposit date: | 2013-10-08 | | Release date: | 2014-08-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.106 Å) | | Cite: | Crystal structure of triple-BRCT-domain of ECT2 and insights into the binding characteristics to CYK-4
Febs Lett., 588, 2014
|
|
8VMC
 
 | |
8W40
 
 | |
4N53
 
 | |
8VM0
 
 | |
4MIU
 
 | |
8W2Y
 
 | |
3GXM
 
 | |
8VG4
 
 | |
8VLP
 
 | |
5WB7
 
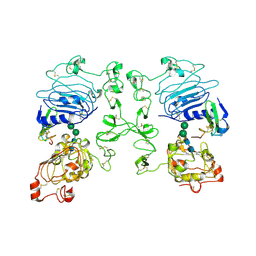 | | Crystal structure of the epidermal growth factor receptor extracellular region in complex with epiregulin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Epidermal growth factor receptor, ... | | Authors: | Freed, D.M, Bessman, N.J, Ferguson, K.M, Lemmon, M.A. | | Deposit date: | 2017-06-28 | | Release date: | 2017-10-18 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.941 Å) | | Cite: | EGFR Ligands Differentially Stabilize Receptor Dimers to Specify Signaling Kinetics.
Cell, 171, 2017
|
|
8VOU
 
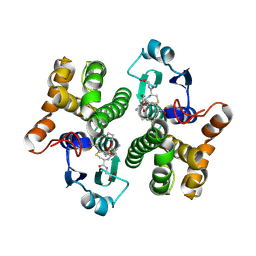 | | Human glutathione transferase M1-1 in complex with the adduct between glutathione and nitrooleic acid | | Descriptor: | GLUTATHIONE, Glutathione S-transferase Mu 1, L-gamma-glutamyl-S-[(8R,9S)-1-carboxy-9-nitroheptadecan-8-yl]-L-cysteinylglycine | | Authors: | Larrieux, N, Steglich, M, Dalla Rizza, J, Buschiazzo, A, Turell, L. | | Deposit date: | 2024-01-16 | | Release date: | 2025-03-12 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Human glutathione transferases catalyze the reaction between glutathione and nitrooleic acid.
J.Biol.Chem., 301, 2025
|
|
4MKY
 
 | |
8W36
 
 | | rabbit actin in the absence of potassium | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Volkmann, N. | | Deposit date: | 2024-02-21 | | Release date: | 2025-02-26 | | Method: | ELECTRON MICROSCOPY (2.27 Å) | | Cite: | rabbit actin in the absence of potassium
To Be Published
|
|
3UBQ
 
 | | Influenza hemagglutinin from the 2009 pandemic in complex with ligand 3SLN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose, ... | | Authors: | Xu, R, Wilson, I.A. | | Deposit date: | 2011-10-24 | | Release date: | 2011-11-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Characterization of the Hemagglutinin Receptor Specificity from the 2009 H1N1 Influenza Pandemic.
J.Virol., 86, 2012
|
|
8W17
 
 | | Lactam bridge synthetic analogue of RgIA | | Descriptor: | Alpha-conotoxin RgIA analogue | | Authors: | Agwa, A.J. | | Deposit date: | 2024-02-15 | | Release date: | 2025-03-05 | | Method: | SOLUTION NMR | | Cite: | On-resin bicyclization improves synthesis of alpha-conotoxin RgIA4-6
To Be Published
|
|
8W4Y
 
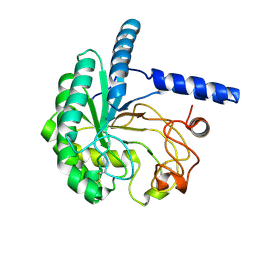 | | Neutron structure of cellulase Cel6A from Phanerochaete chrysosporium at room temperature, low-D2O-solvent | | Descriptor: | Glucanase | | Authors: | Tachioka, M, Yamaguchi, S, Nakamura, A, Ishida, T, Kusaka, K, Yamada, T, Yano, N, Chatake, T, Tamada, T, Takeda, K, Niwa, S, Tanaka, H, Takahashi, S, Inaka, K, Furubayashi, N, Deguchi, S, Samejima, M, Igarashi, K. | | Deposit date: | 2023-08-25 | | Release date: | 2025-03-12 | | Method: | NEUTRON DIFFRACTION (1.4 Å), X-RAY DIFFRACTION | | Cite: | Deprotonated Arginine Controls a Putative Catalytic Base in Invert-ing Family 6 Glycoside Hydrolase
To Be Published
|
|
4V6E
 
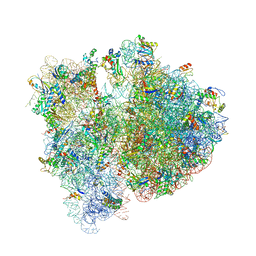 | | Crystal structure of the E. coli 70S ribosome in an intermediate state of ratcheting | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Zhang, W, Dunkle, J.A, Cate, J.H.D. | | Deposit date: | 2009-06-28 | | Release date: | 2014-07-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.712 Å) | | Cite: | Structures of the ribosome in intermediate States of ratcheting.
Science, 325, 2009
|
|
8VGX
 
 | |
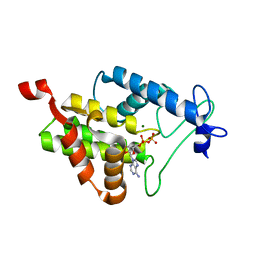
4N67
 
 | |
8W4W
 
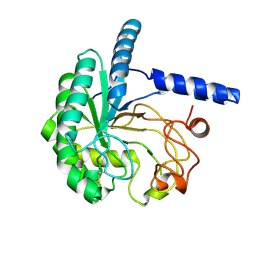 | | Neutron structure of cellulase Cel6A from Phanerochaete chrysosporium at room temperature | | Descriptor: | Glucanase | | Authors: | Tachioka, M, Yamaguchi, S, Nakamura, A, Ishida, T, Kusaka, K, Yamada, T, Yano, N, Chatake, T, Tamada, T, Takeda, K, Niwa, S, Tanaka, H, Takahashi, S, Inaka, K, Furubayashi, N, Deguchi, S, Samejima, M, Igarashi, K. | | Deposit date: | 2023-08-25 | | Release date: | 2025-03-12 | | Method: | NEUTRON DIFFRACTION (1.36 Å), X-RAY DIFFRACTION | | Cite: | Deprotonated Arginine Controls a Putative Catalytic Base in Invert-ing Family 6 Glycoside Hydrolase
To Be Published
|
|
4N6C
 
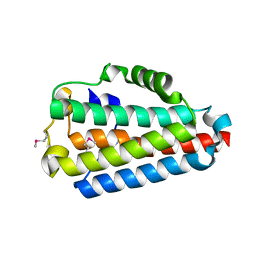 | | Crystal Structure of the B1RZQ2 protein from Streptococcus pneumoniae. Northeast Structural Genomics Consortium (NESG) Target SpR36. | | Descriptor: | BROMIDE ION, uncharacterized protein | | Authors: | Vorobiev, S, Seetharaman, J, Patel, D, Xiao, R, Ciccosanti, C, Wang, D, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-10-11 | | Release date: | 2013-10-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.548 Å) | | Cite: | Crystal Structure of the B1RZQ2 protein from Streptococcus pneumoniae.
To be Published
|
|
4V8H
 
 | | Crystal structure of HPF bound to the 70S ribosome. | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S Ribosomal Protein S10, ... | | Authors: | Polikanov, Y.S, Blaha, G.M, Steitz, T.A. | | Deposit date: | 2011-12-11 | | Release date: | 2014-07-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | How Hibernation Factors RMF, HPF, and YfiA Turn Off Protein Synthesis.
Science, 336, 2012
|
|
8VV1
 
 | | Estrogen receptor alpha ligand binding domain in complex with palazestrant | | Descriptor: | Estrogen receptor, GLYCEROL, palazestrant | | Authors: | Ng, R.A, Barratt, S, Parisian, A, Palanisamy, G, Sun, R, Robello, B, Pena, G, Sapugay, J, Yeghikyan, D, Duncan, A, Andersen, S.E, Chawla, R, Rich, B, Hearn, B, Harmon, C, Hodges-Gallagher, L, Kushner, P.J, Greene, G.L, Myles, D.C, Fanning, S.W. | | Deposit date: | 2024-01-30 | | Release date: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.196 Å) | | Cite: | Estrogen receptor alpha ligand binding domain in complex with palazestrant
To Be Published
|
|
8VLD
 
 | |
