2BF7
 
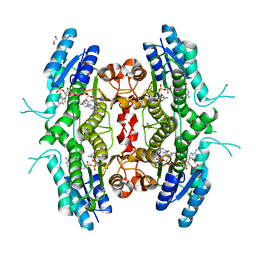 | | Leishmania major pteridine reductase 1 in complex with NADP and biopterin | | Descriptor: | 1,2-ETHANEDIOL, 7,8-DIHYDROBIOPTERIN, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Schuettelkopf, A.W, Hunter, W.N. | | Deposit date: | 2004-12-06 | | Release date: | 2005-08-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of Leishmania Major Pteridine Reductase Complexes Reveal the Active Site Features Important for Ligand Binding and to Guide Inhibitor Design
J.Mol.Biol., 352, 2005
|
|
2BFA
 
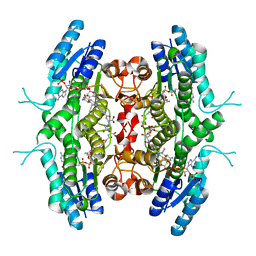 | | Leishmania major pteridine reductase 1 in complex with NADP and CB3717 | | Descriptor: | 1,2-ETHANEDIOL, 10-PROPARGYL-5,8-DIDEAZAFOLIC ACID, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Schuettelkopf, A.W, Hunter, W.N. | | Deposit date: | 2004-12-06 | | Release date: | 2005-08-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structures of Leishmania Major Pteridine Reductase Complexes Reveal the Active Site Features Important for Ligand Binding and to Guide Inhibitor Design
J.Mol.Biol., 352, 2005
|
|
2BFP
 
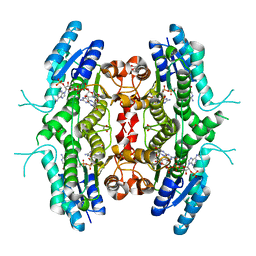 | | Leishmania major pteridine reductase 1 in complex with NADP and tetrahydrobiopterin | | Descriptor: | 1,2-ETHANEDIOL, 5,6,7,8-TETRAHYDROBIOPTERIN, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Schuettelkopf, A.W, Hunter, W.N. | | Deposit date: | 2004-12-10 | | Release date: | 2005-08-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structures of Leishmania Major Pteridine Reductase Complexes Reveal the Active Site Features Important for Ligand Binding and to Guide Inhibitor Design
J.Mol.Biol., 352, 2005
|
|
5XN2
 
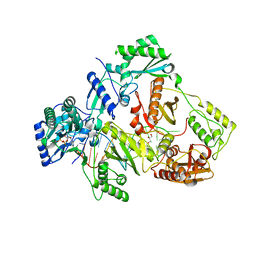 | | HIV-1 reverse transcriptase Q151M:DNA:dGTP ternary complex | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, 38-MER DNA aptamer, GLYCEROL, ... | | Authors: | Yasutake, Y, Tamura, N, Hayashi, H, Maeda, K. | | Deposit date: | 2017-05-17 | | Release date: | 2018-02-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.381 Å) | | Cite: | HIV-1 with HBV-associated Q151M substitution in RT becomes highly susceptible to entecavir: structural insights into HBV-RT inhibition by entecavir.
Sci Rep, 8, 2018
|
|
2QX0
 
 | | Crystal Structure of Yersinia pestis HPPK (Ternary Complex) | | Descriptor: | 2-AMINO-6-HYDROXYMETHYL-7,8-DIHYDRO-3H-PTERIDIN-4-ONE, 7,8-dihydro-6-hydroxymethylpterin-pyrophosphokinase, DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, ... | | Authors: | Blaszczyk, J, Cherry, S, Tropea, J.E, Waugh, D.S, Ji, X. | | Deposit date: | 2007-08-10 | | Release date: | 2007-10-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and activity of Yersinia pestis 6-hydroxymethyl-7,8-dihydropterin pyrophosphokinase as a novel target for the development of antiplague therapeutics.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
8BMR
 
 | | Cryo-EM structure of the wild-type solitary ECF module in MSP2N2 lipid nanodiscs in the ATPase open and nucleotide-free conformation | | Descriptor: | Energy-coupling factor transporter ATP-binding protein EcfA1, Energy-coupling factor transporter ATP-binding protein EcfA2, Energy-coupling factor transporter transmembrane protein EcfT | | Authors: | Thangaratnarajah, C, Rheinberger, J, Paulino, C, Slotboom, D.J. | | Deposit date: | 2022-11-10 | | Release date: | 2023-08-02 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Expulsion mechanism of the substrate-translocating subunit in ECF transporters.
Nat Commun, 14, 2023
|
|
3GN1
 
 | | Structure of Pteridine Reductase 1 (PTR1) from TRYPANOSOMA BRUCEI in ternary complex with cofactor (NADP+) and inhibitor (DDD00067116) | | Descriptor: | 1H-benzimidazol-2-amine, ACETATE ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Tulloch, L.B, Brenk, R, Hunter, W.N. | | Deposit date: | 2009-03-16 | | Release date: | 2009-12-29 | | Last modified: | 2011-09-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | One scaffold, three binding modes: novel and selective pteridine reductase 1 inhibitors derived from fragment hits discovered by virtual screening.
J.Med.Chem., 52, 2009
|
|
5H1T
 
 | | Complex structure of TRIM24 PHD-bromodomain and inhibitor 1 | | Descriptor: | DIMETHYL SULFOXIDE, Transcription intermediary factor 1-alpha, ZINC ION, ... | | Authors: | Liu, J. | | Deposit date: | 2016-10-11 | | Release date: | 2017-02-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.951 Å) | | Cite: | The polar warhead of a TRIM24 bromodomain inhibitor rearranges a water-mediated interaction network
FEBS J., 284, 2017
|
|
1OUD
 
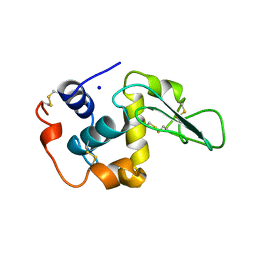 | | CONTRIBUTION OF HYDROPHOBIC RESIDUES TO THE STABILITY OF HUMAN LYSOZYME: X-RAY STRUCTURE OF THE V121A MUTANT | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Takano, K, Yamagata, Y, Fujii, S, Yutani, K. | | Deposit date: | 1996-08-23 | | Release date: | 1997-02-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Contribution of the hydrophobic effect to the stability of human lysozyme: calorimetric studies and X-ray structural analyses of the nine valine to alanine mutants.
Biochemistry, 36, 1997
|
|
3LU8
 
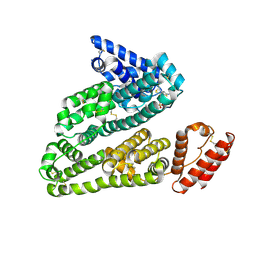 | | Human serum albumin in complex with compound 3 | | Descriptor: | N-[5-(5-{[(2,4-dimethyl-1,3-thiazol-5-yl)sulfonyl]amino}-6-fluoropyridin-3-yl)-4-methyl-1,3-thiazol-2-yl]acetamide, Serum albumin | | Authors: | Buttar, D, Colclough, N, Gerhardt, S, MacFaul, P.A, Phillips, S.D, Plowright, A, Whittamore, P, Tam, K, Maskos, K, Steinbacher, S, Steuber, H. | | Deposit date: | 2010-02-17 | | Release date: | 2010-10-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A combined spectroscopic and crystallographic approach to probing drug-human serum albumin interactions
Bioorg.Med.Chem., 18, 2010
|
|
5TVP
 
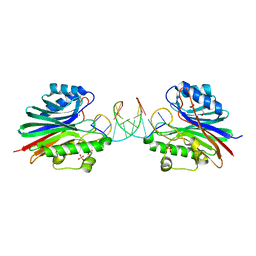 | |
5MVP
 
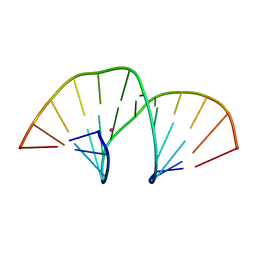 | | Crystal structure of an A-DNA dodecamer containing the GGGCCC motif | | Descriptor: | DNA, POTASSIUM ION | | Authors: | Hardwick, J.S, Ptchelkine, D, Phillips, S.E.V, Brown, T. | | Deposit date: | 2017-01-17 | | Release date: | 2017-05-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.606 Å) | | Cite: | 5-Formylcytosine does not change the global structure of DNA.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5MYJ
 
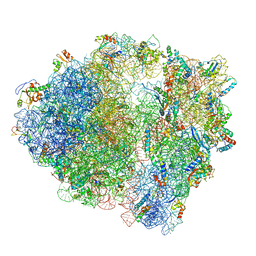 | | Structure of 70S ribosome from Lactococcus lactis | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Franken, L.E, Oostergetel, G.T, Pijning, T, Puri, P, Boekema, E.J, Poolman, B, Guskov, A. | | Deposit date: | 2017-01-26 | | Release date: | 2017-10-11 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (5.6 Å) | | Cite: | A general mechanism of ribosome dimerization revealed by single-particle cryo-electron microscopy.
Nat Commun, 8, 2017
|
|
5MBA
 
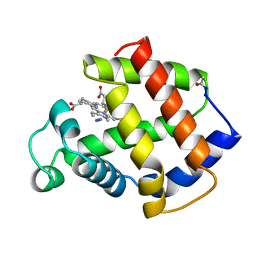 | | BINDING MODE OF AZIDE TO FERRIC APLYSIA LIMACINA MYOGLOBIN. CRYSTALLOGRAPHIC ANALYSIS AT 1.9 ANGSTROMS RESOLUTION | | Descriptor: | AZIDE ION, MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Bolognesi, M, Onesti, S, Gatti, G, Coda, A, Ascenzi, P, Brunori, M. | | Deposit date: | 1991-01-14 | | Release date: | 1992-07-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Binding mode of azide to ferric Aplysia limacina myoglobin. Crystallographic analysis at 1.9 A resolution.
J.Mol.Recog., 4, 1991
|
|
3LOY
 
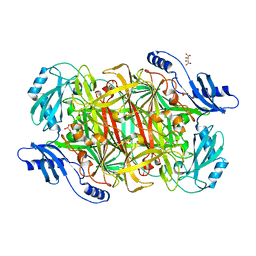 | | Crystal structure of a Copper-containing benzylamine oxidase from Hansenula Polymorpha | | Descriptor: | COPPER (II) ION, GLYCEROL, L(+)-TARTARIC ACID, ... | | Authors: | Klema, V.J, Johnson, B.J, Wilmot, C.M. | | Deposit date: | 2010-02-04 | | Release date: | 2010-03-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Kinetic and structural analysis of substrate specificity in two copper amine oxidases from Hansenula polymorpha.
Biochemistry, 49, 2010
|
|
5TUV
 
 | |
1OUG
 
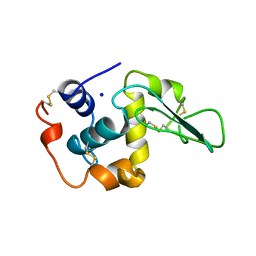 | | CONTRIBUTION OF HYDROPHOBIC RESIDUES TO THE STABILITY OF HUMAN LYSOZYME: X-RAY STRUCTURE OF THE V2A MUTANT | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Takano, K, Yamagata, Y, Fujii, S, Yutani, K. | | Deposit date: | 1996-08-23 | | Release date: | 1997-02-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Contribution of the hydrophobic effect to the stability of human lysozyme: calorimetric studies and X-ray structural analyses of the nine valine to alanine mutants.
Biochemistry, 36, 1997
|
|
3LU7
 
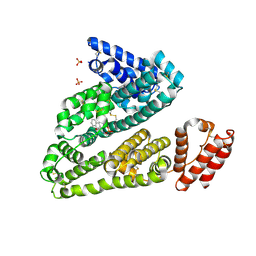 | | Human serum albumin in complex with compound 2 | | Descriptor: | 4-[(1R,2R)-2-{[(5-fluoro-1H-indol-2-yl)carbonyl]amino}-2,3-dihydro-1H-inden-1-yl]butanoic acid, PHOSPHATE ION, Serum albumin | | Authors: | Buttar, D, Colclough, N, Gerhardt, S, MacFaul, P.A, Phillips, S.D, Plowright, A, Whittamore, P, Tam, K, Maskos, K, Steinbacher, S, Steuber, H. | | Deposit date: | 2010-02-17 | | Release date: | 2010-10-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A combined spectroscopic and crystallographic approach to probing drug-human serum albumin interactions
Bioorg.Med.Chem., 18, 2010
|
|
6RQG
 
 | |
2CAU
 
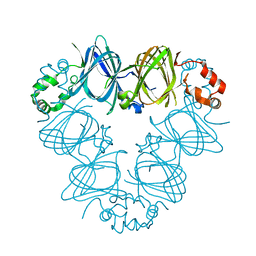 | | CANAVALIN FROM JACK BEAN | | Descriptor: | PROTEIN (CANAVALIN) | | Authors: | Ko, T.-P, Day, J, Macpherson, A. | | Deposit date: | 1998-11-20 | | Release date: | 1998-11-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The refined structure of canavalin from jack bean in two crystal forms at 2.1 and 2.0 A resolution.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
5GIJ
 
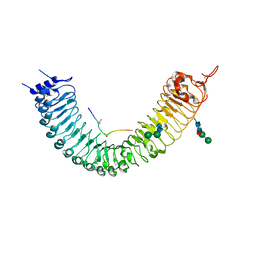 | | Crystal structure of TDR-TDIF complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Leucine-rich repeat receptor-like protein kinase TDR, ... | | Authors: | Morita, J, Kato, K, Ishitani, R, Nishimasu, H, Nureki, O. | | Deposit date: | 2016-06-23 | | Release date: | 2016-08-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of the plant receptor-like kinase TDR in complex with the TDIF peptide
Nat Commun, 7, 2016
|
|
1P32
 
 | | CRYSTAL STRUCTURE OF HUMAN P32, A DOUGHNUT-SHAPED ACIDIC MITOCHONDRIAL MATRIX PROTEIN | | Descriptor: | MITOCHONDRIAL MATRIX PROTEIN, SF2P32 | | Authors: | Jiang, J, Zhang, Y, Krainer, A.R, Xu, R.-M. | | Deposit date: | 1998-11-02 | | Release date: | 1999-04-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of human p32, a doughnut-shaped acidic mitochondrial matrix protein.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1OQN
 
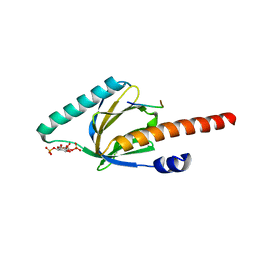 | | Crystal structure of the phosphotyrosine binding domain (PTB) of mouse Disabled 1 (Dab1) | | Descriptor: | Alzheimer's disease amyloid A4 protein homolog, D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, Disabled homolog 1 | | Authors: | Yun, M, Keshvara, L, Park, C.-G, Zhang, Y.-M, Dickerson, J.B, Zheng, J, Rock, C.O, Curran, T, Park, H.-W. | | Deposit date: | 2003-03-10 | | Release date: | 2003-08-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of the Dab homology domains of mouse disabled 1 and 2
J.Biol.Chem., 278, 2003
|
|
1ORH
 
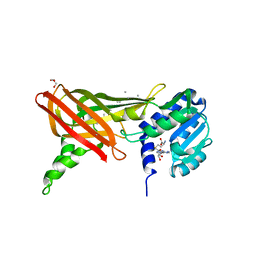 | | Structure of the Predominant Protein Arginine Methyltransferase PRMT1 | | Descriptor: | GLYCEROL, Protein arginine N-methyltransferase 1, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Zhang, X, Cheng, X. | | Deposit date: | 2003-03-13 | | Release date: | 2003-08-26 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Structure of the Predominant Protein Arginine Methyltransferase PRMT1 and Analysis of Its Binding to Substrate Peptides
Structure, 11, 2003
|
|
6RNW
 
 | |
