8TOW
 
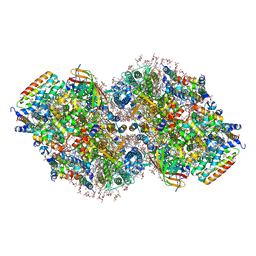 | | Structure of a mutated photosystem II complex reveals perturbation of the oxygen-evolving complex | | Descriptor: | (3R)-beta,beta-caroten-3-ol, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Flesher, D.A, Liu, J, Wang, J, Gisriel, C.J, Yang, K.R, Batista, V.S, Debus, R.J, Brudvig, G.W. | | Deposit date: | 2023-08-04 | | Release date: | 2024-06-19 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.14 Å) | | Cite: | Mutation-induced shift of the photosystem II active site reveals insight into conserved water channels.
J.Biol.Chem., 300, 2024
|
|
6NCW
 
 | |
5LB6
 
 | |
5LBE
 
 | |
7N03
 
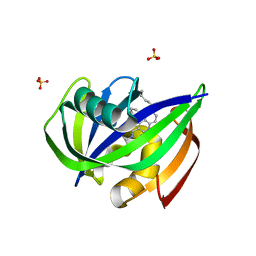 | | Crystal structure of MTH1 in complex with compound 31 | | Descriptor: | 4-anilino-6-(hexylamino)-N-methylquinoline-3-carboxamide, 7,8-dihydro-8-oxoguanine triphosphatase, SULFATE ION | | Authors: | Eron, S.J. | | Deposit date: | 2021-05-24 | | Release date: | 2021-11-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Development of an AchillesTAG degradation system and its application to control CAR-T activity
Curr Res Chem Biol, 1, 2021
|
|
9BWI
 
 | | Crystal structure of cellulose oxidative enzyme in acidic pH with glycerol | | Descriptor: | COPPER (II) ION, Cellulose oxidative enzyme, GLYCEROL | | Authors: | Morais, M.A.B, Santos, C.A, Araujo, E.A, Santos, C.R, Morao, L.G, Motta, M.L, Murakami, M.T. | | Deposit date: | 2024-05-21 | | Release date: | 2024-12-11 | | Last modified: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A metagenomic 'dark matter' enzyme catalyses oxidative cellulose conversion.
Nature, 639, 2025
|
|
5LBZ
 
 | | Structure of the human quinone reductase 2 (NQO2) in complex with pacritinib | | Descriptor: | 11-(2-pyrrolidin-1-yl-ethoxy)-14,19-dioxa-5,7,26-triaza-tetracyclo[19.3.1.1(2,6).1(8,12)]heptacosa-1(25),2(26),3,5,8,10,12(27),16,21,23-decaene, FLAVIN-ADENINE DINUCLEOTIDE, Ribosyldihydronicotinamide dehydrogenase [quinone], ... | | Authors: | Schneider, S, Medard, G, Kuster, B. | | Deposit date: | 2016-06-17 | | Release date: | 2017-11-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The target landscape of clinical kinase drugs.
Science, 358, 2017
|
|
6NCI
 
 | | Crystal structure of CDP-Chase: Vector data collection | | Descriptor: | D-ribose, DI(HYDROXYETHYL)ETHER, PHOSPHATE ION, ... | | Authors: | Miller, M.S, Shi, W, Gabelli, S.B. | | Deposit date: | 2018-12-11 | | Release date: | 2019-02-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Getting the Most Out of Your Crystals: Data Collection at the New High-Flux, Microfocus MX Beamlines at NSLS-II.
Molecules, 24, 2019
|
|
7N13
 
 | | Crystal structure of MTH1 in complex with compound 32 | | Descriptor: | 4-anilino-6-[4-(butylcarbamoyl)-3-fluorophenyl]-N-cyclopropyl-7-fluoroquinoline-3-carboxamide, 7,8-dihydro-8-oxoguanine triphosphatase, SULFATE ION | | Authors: | Eron, S.J. | | Deposit date: | 2021-05-26 | | Release date: | 2021-11-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Development of an AchillesTAG degradation system and its application to control CAR-T activity
Curr Res Chem Biol, 1, 2021
|
|
5KZW
 
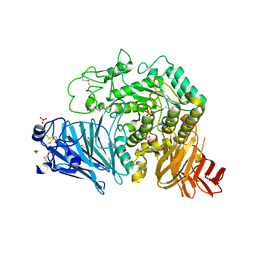 | | Crystal structure of human GAA | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Deming, D.T, Garman, S.C. | | Deposit date: | 2016-07-25 | | Release date: | 2017-07-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structure of human GAA: structural basis of Pompe disease
To be published
|
|
9C4E
 
 | |
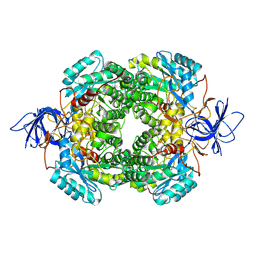
9C86
 
 | |
8TBA
 
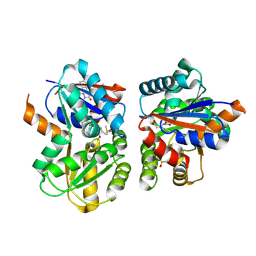 | |
4LDY
 
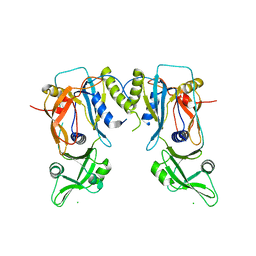 | | Crystal structure of the DNA binding domain of the G245A mutant of arabidopsis thaliana auxin reponse factor 1 | | Descriptor: | Auxin response factor 1, CHLORIDE ION | | Authors: | Boer, D.R, Freire-Rios, A, van den Berg, W.M.A, Weijers, D, Coll, M. | | Deposit date: | 2013-06-25 | | Release date: | 2014-02-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis for DNA Binding Specificity by the Auxin-Dependent ARF Transcription Factors.
Cell(Cambridge,Mass.), 156, 2014
|
|
6NI3
 
 | | B2V2R-Gs protein subcomplex of a GPCR-G protein-beta-arrestin mega-complex | | Descriptor: | 8-[(1R)-2-{[1,1-dimethyl-2-(2-methylphenyl)ethyl]amino}-1-hydroxyethyl]-5-hydroxy-2H-1,4-benzoxazin-3(4H)-one, Endolysin,Beta-2 adrenergic receptor chimera, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Nguyen, A.H, Thomsen, A.R.B, Cahill, T.J, des Georges, A, Lefkowitz, R.J. | | Deposit date: | 2018-12-26 | | Release date: | 2019-11-20 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of an endosomal signaling GPCR-G protein-beta-arrestin megacomplex.
Nat.Struct.Mol.Biol., 26, 2019
|
|
5KLK
 
 | | Crystal structure of 2-aminomuconate 6-semialdehyde dehydrogenase N169D in complex with NAD+ and 2-hydroxymuconate-6-semialdehyde | | Descriptor: | (2E,4E)-2-hydroxy-6-oxohexa-2,4-dienoic acid, 2-aminomuconate 6-semialdehyde dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Yang, Y, Davis, I, Ha, U, Wang, Y, Shin, I, Liu, A. | | Deposit date: | 2016-06-24 | | Release date: | 2016-11-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.006 Å) | | Cite: | A Pitcher-and-Catcher Mechanism Drives Endogenous Substrate Isomerization by a Dehydrogenase in Kynurenine Metabolism.
J.Biol.Chem., 291, 2016
|
|
9BZ0
 
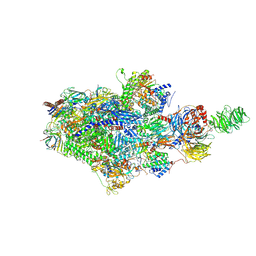 | | Structure of an STK19-containing TC-NER complex | | Descriptor: | DET1- and DDB1-associated protein 1, DNA (35-MER), DNA (49-MER), ... | | Authors: | Mevissen, T.E.T, Kuemmecke, M, Farnung, L, Walter, J.C. | | Deposit date: | 2024-05-24 | | Release date: | 2024-12-25 | | Last modified: | 2025-01-15 | | Method: | ELECTRON MICROSCOPY (1.9 Å) | | Cite: | STK19 positions TFIIH for cell-free transcription-coupled DNA repair.
Cell, 187, 2024
|
|
5L94
 
 | |
5LAG
 
 | |
8U19
 
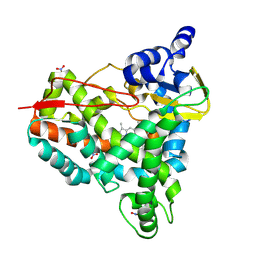 | | Crystal structure of SyoA bound to syringol | | Descriptor: | 2,6-dimethoxyphenol, Cytochrome P450, MAGNESIUM ION, ... | | Authors: | Harlington, A.C, Shearwin, K.E, Bell, S.G, Whelan, F. | | Deposit date: | 2023-08-31 | | Release date: | 2024-07-24 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Structural insights into S-lignin O-demethylation via a rare class of heme peroxygenase enzymes.
Nat Commun, 16, 2025
|
|
9CGP
 
 | |
5LBT
 
 | | Structure of the human quinone reductase 2 (NQO2) in complex with imiquimod | | Descriptor: | 1-(2-methylpropyl)imidazo[4,5-c]quinolin-4-amine, FLAVIN-ADENINE DINUCLEOTIDE, Ribosyldihydronicotinamide dehydrogenase [quinone], ... | | Authors: | Schneider, S, Gross, O. | | Deposit date: | 2016-06-17 | | Release date: | 2016-07-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Imiquimod Inhibits Mitochondrial Complex I and Induces K+ efflux-independent Nlrp3 Inflammasome Activation via Nek7
To Be Published
|
|
5LC5
 
 | | Structure of mammalian respiratory Complex I, class2 | | Descriptor: | Acyl carrier protein, mitochondrial, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Vinothkumar, K.R, Zhu, J, Hirst, J. | | Deposit date: | 2016-06-19 | | Release date: | 2016-09-07 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.35 Å) | | Cite: | Structure of mammalian respiratory complex I.
Nature, 536, 2016
|
|
8U09
 
 | | Crystal structure of substrate-free SyoA | | Descriptor: | ACETATE ION, Cytochrome P450, GLYCEROL, ... | | Authors: | Harlington, A.C, Shearwin, K.E, Bell, S.G, Whelan, F. | | Deposit date: | 2023-08-28 | | Release date: | 2024-07-24 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural insights into S-lignin O-demethylation via a rare class of heme peroxygenase enzymes.
Nat Commun, 16, 2025
|
|
9CL1
 
 | | Particulate methane monooxygenase in 0.02% DDM | | Descriptor: | COPPER (II) ION, Methane monooxygenase, C subunit, ... | | Authors: | Tucci, F.J, Rosenzweig, A.C. | | Deposit date: | 2024-07-10 | | Release date: | 2025-01-08 | | Last modified: | 2025-01-15 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Structures of methane and ammonia monooxygenases in native membranes.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
