1EE3
 
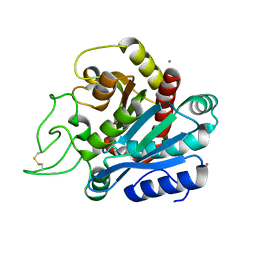 | | Cadmium-substituted bovine pancreatic carboxypeptidase A (alfa-form) at pH 7.5 and 2 mM chloride in monoclinic crystal form | | Descriptor: | CADMIUM ION, PROTEIN (CARBOXYPEPTIDASE A) | | Authors: | Jensen, F, Bukrinsky, T, Bjerrum, J, Larsen, S. | | Deposit date: | 2000-01-30 | | Release date: | 2002-06-26 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Three high-resolution crystal structures of cadmium-substituted carboxypeptidase A provide insight into the enzymatic function
J.BIOL.INORG.CHEM., 7, 2002
|
|
6HH1
 
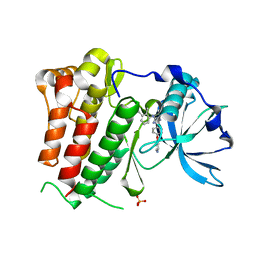 | | Structure of c-Kit with allosteric inhibitor 3G8 | | Descriptor: | Mast/stem cell growth factor receptor Kit,Mast/stem cell growth factor receptor Kit, PHOSPHATE ION, ~{N}-(2,3-dimethylphenyl)-5-(4-pyridin-4-yloxyphenyl)-4~{H}-1,2,4-triazol-3-amine | | Authors: | Wrasidlo, W.J, Cheresh, D.A. | | Deposit date: | 2018-08-24 | | Release date: | 2019-09-18 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure of c-Kit with allosteric inhibitor 3G8
To Be Published
|
|
1EM8
 
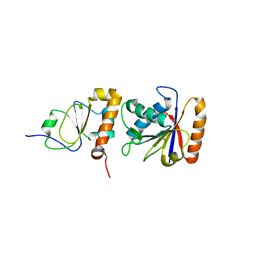 | | Crystal structure of chi and psi subunit heterodimer from DNA POL III | | Descriptor: | DNA POLYMERASE III CHI SUBUNIT, DNA POLYMERASE III PSI SUBUNIT | | Authors: | Gulbis, J.M, Finkelstein, J, O'Donnell, M, Kuriyan, J. | | Deposit date: | 2000-03-16 | | Release date: | 2003-08-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the chi:psi sub-assembly of the Escherichia coli DNA polymerase clamp-loader complex.
Eur.J.Biochem., 271, 2004
|
|
5NUM
 
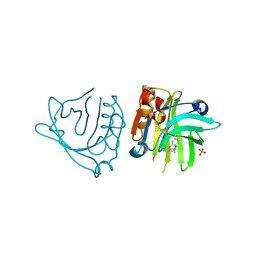 | | Engineered beta-lactoglobulin: variant F105L-L39A in complex with chlorpromazine (LG-LA-CLP) | | Descriptor: | 3-(2-chloro-10H-phenothiazin-10-yl)-N,N-dimethylpropan-1-amine, Beta-lactoglobulin, PHOSPHATE ION | | Authors: | Loch, J.I, Bonarek, P, Tworzydlo, M, Lazinska, I, Szydlowska, J, Lewinski, K. | | Deposit date: | 2017-04-30 | | Release date: | 2018-04-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The engineered beta-lactoglobulin with complementarity to the chlorpromazine chiral conformers.
Int. J. Biol. Macromol., 114, 2018
|
|
6HMZ
 
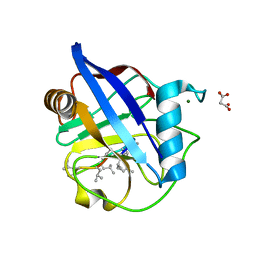 | | Crystal Structure of a Single-Domain Cyclophilin from Brassica napus Phloem Sap | | Descriptor: | Cyclosporin, MAGNESIUM ION, MALONATE ION, ... | | Authors: | Falke, S, Hanhart, P, Garbe, M, Thiess, M, Betzel, C, Kehr, J. | | Deposit date: | 2018-09-13 | | Release date: | 2018-11-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Enzyme activity and structural features of three single-domain phloem cyclophilins from Brassica napus.
Sci Rep, 9, 2019
|
|
5NXA
 
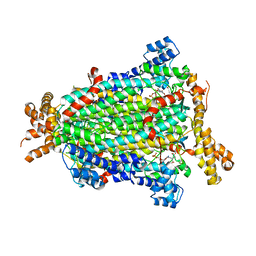 | | Crystal structure of Neanderthal Adenylosuccinate Lyase (ADSL)in complex with its products AICAR and fumarate | | Descriptor: | AMINOIMIDAZOLE 4-CARBOXAMIDE RIBONUCLEOTIDE, Adenylosuccinate lyase, CHLORIDE ION, ... | | Authors: | Van Laer, B, Kapp, U, Soler-Lopez, M, Leonard, G, Mueller-Dieckmann, C. | | Deposit date: | 2017-05-09 | | Release date: | 2018-05-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular comparison of Neanderthal and Modern Human adenylosuccinate lyase.
Sci Rep, 8, 2018
|
|
1EI9
 
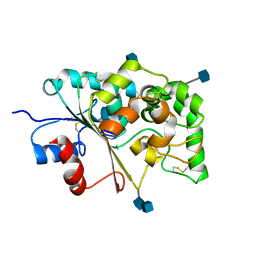 | | CRYSTAL STRUCTURE OF PALMITOYL PROTEIN THIOESTERASE 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, PALMITOYL PROTEIN THIOESTERASE 1 | | Authors: | Bellizzi III, J.J, Widom, J, Kemp, C, Lu, J.Y, Das, A.K, Hofmann, S.L, Clardy, J. | | Deposit date: | 2000-02-24 | | Release date: | 2000-04-26 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The crystal structure of palmitoyl protein thioesterase 1 and the molecular basis of infantile neuronal ceroid lipofuscinosis.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
5NYB
 
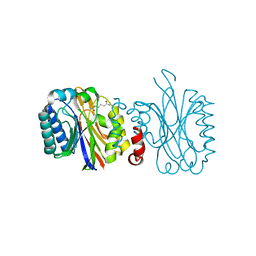 | |
5U5N
 
 | |
5ITS
 
 | | Crystal structure of LOG from Corynebacterium glutamicum | | Descriptor: | Cytokinin riboside 5'-monophosphate phosphoribohydrolase, GLYCEROL, PHOSPHATE ION | | Authors: | Seo, H.-G, Kim, K.-J. | | Deposit date: | 2016-03-17 | | Release date: | 2016-08-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for cytokinin production by LOG from Corynebacterium glutamicum
Sci Rep, 6, 2016
|
|
1ELL
 
 | | CADMIUM-SUBSTITUTED BOVINE PANCREATIC CARBOXYPEPTIDASE A (ALFA-FORM) AT PH 7.5 AND 0.25 M CHLORIDE IN MONOCLINIC CRYSTAL FORM. | | Descriptor: | CADMIUM ION, CARBOXYPEPTIDASE A | | Authors: | Jensen, F, Bukrinsky, T, Bjerrum, J, Larsen, S. | | Deposit date: | 2000-03-14 | | Release date: | 2002-06-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Three high-resolution crystal structures of cadmium-substituted carboxypeptidase A provide insight into the enzymatic function
J.BIOL.INORG.CHEM., 7, 2002
|
|
6HKI
 
 | |
5NVG
 
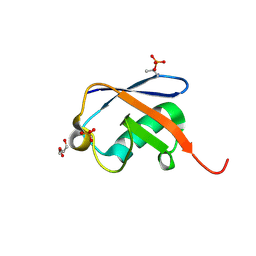 | | Thr12 Phosphorylated Ubiquitin | | Descriptor: | DI(HYDROXYETHYL)ETHER, PHOSPHATE ION, Polyubiquitin-B | | Authors: | Huguenin-Dezot, N. | | Deposit date: | 2017-05-04 | | Release date: | 2017-05-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | Biosynthesis and genetic encoding of phosphothreonine through parallel selection and deep sequencing.
Nat. Methods, 14, 2017
|
|
5U7L
 
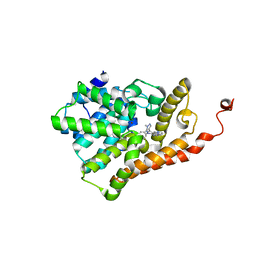 | | PDE2 catalytic domain complexed with inhibitors | | Descriptor: | (3R)-1-{3-[5-(4-ethylphenyl)-1-methyl-1H-pyrazol-4-yl]-1-methyl-1H-pyrazolo[3,4-d]pyrimidin-4-yl}-N,N-dimethylpyrrolidin-3-amine, MAGNESIUM ION, ZINC ION, ... | | Authors: | Pandit, J, Parris, K. | | Deposit date: | 2016-12-12 | | Release date: | 2017-06-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Application of Structure-Based Design and Parallel Chemistry to Identify a Potent, Selective, and Brain Penetrant Phosphodiesterase 2A Inhibitor.
J. Med. Chem., 60, 2017
|
|
5NWY
 
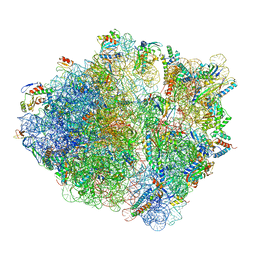 | | 2.9 A cryo-EM structure of VemP-stalled ribosome-nascent chain complex | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Su, T, Cheng, J, Sohmen, D, Hedman, R, Berninghausen, O, von Heijne, G, Wilson, D.N, Beckmann, R. | | Deposit date: | 2017-05-08 | | Release date: | 2017-07-19 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | The force-sensing peptide VemP employs extreme compaction and secondary structure formation to induce ribosomal stalling.
Elife, 6, 2017
|
|
6HSN
 
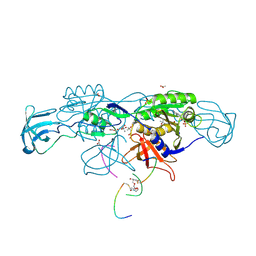 | | Crystal structure of the ternary complex of GephE-ADP-GABA(A) receptor derived peptide | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,1'-[ethane-1,2-diylbis(oxyethane-2,1-diyl)]bis(1H-pyrrole-2,5-dione), ACETATE ION, ... | | Authors: | Kasaragod, V.B, Schindelin, H. | | Deposit date: | 2018-10-01 | | Release date: | 2019-01-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Elucidating the Molecular Basis for Inhibitory Neurotransmission Regulation by Artemisinins.
Neuron, 101, 2019
|
|
5TW3
 
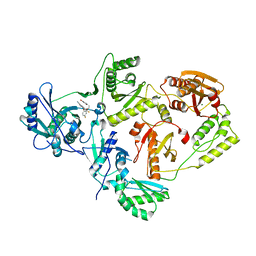 | | Crystal Structure of HIV-1 Reverse Transcriptase in Complex with 5-(2-(2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy)-4-fluorophenoxy)-7-fluoro-2-naphthonitrile (JLJ636), a Non-nucleoside Inhibitor | | Descriptor: | 5-{2-[2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy]-4-fluorophenoxy}-7-fluoronaphthalene-2-carbonitrile, HIV-1 REVERSE TRANSCRIPTASE, P51 SUBUNIT, ... | | Authors: | Chan, A.H, Anderson, K.S. | | Deposit date: | 2016-11-11 | | Release date: | 2017-03-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.853 Å) | | Cite: | Structural and Preclinical Studies of Computationally Designed Non-Nucleoside Reverse Transcriptase Inhibitors for Treating HIV infection.
Mol. Pharmacol., 91, 2017
|
|
5TPV
 
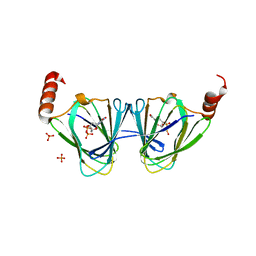 | | X-ray structure of WlaRA (TDP-fucose-3,4-ketoisomerase) from Campylobacter jejuni | | Descriptor: | PHOSPHATE ION, THYMIDINE-5'-DIPHOSPHATE, WlaRA, ... | | Authors: | Holden, H.M, Thoden, J.B, Li, Z.A, Riegert, A.S, Goneau, M.-F, Cunningham, A.M, Vinograd, E, Schoenhofen, I.C, Gilbert, M, Li, J. | | Deposit date: | 2016-10-21 | | Release date: | 2017-02-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Characterization of the dTDP-Fuc3N and dTDP-Qui3N biosynthetic pathways in Campylobacter jejuni 81116.
Glycobiology, 27, 2017
|
|
5NWX
 
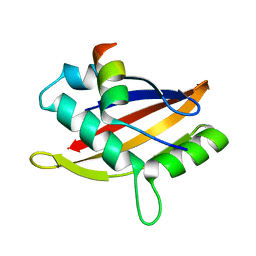 | | Insight into the molecular recognition mechanism of the coactivator NCoA1 by STAT6 | | Descriptor: | Nuclear receptor coactivator 1, Signal transducer and activator of transcription 6 | | Authors: | Russo, L, Giller, K, Pfitzner, E, Griesinger, C, Becker, S. | | Deposit date: | 2017-05-08 | | Release date: | 2017-12-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Insight into the molecular recognition mechanism of the coactivator NCoA1 by STAT6.
Sci Rep, 7, 2017
|
|
6HND
 
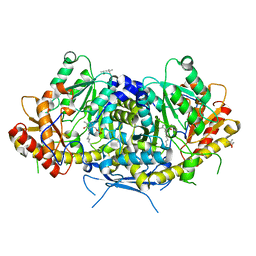 | | Crystal structure of the aromatic aminotransferase Aro9 from C. Albicans | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Aromatic-amino-acid:2-oxoglutarate transaminase, POTASSIUM ION, ... | | Authors: | Kiliszek, A, Rzad, K, Rypniewski, W, Milewski, S, Gabriel, I. | | Deposit date: | 2018-09-14 | | Release date: | 2019-02-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Crystal structures of aminotransferases Aro8 and Aro9 from Candida albicans and structural insights into their properties.
J.Struct.Biol., 205, 2019
|
|
5O36
 
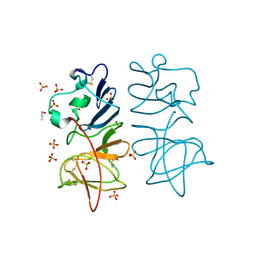 | | Japanese encephalitis virus non-structural protein 1' C-terminal domain | | Descriptor: | Japanese encephalitis virus non-structural protein 1' (NS1'),Japanese encephalitis virus non-structural protein 1' (NS1'), N-PROPANOL, SULFATE ION | | Authors: | Thanalai, P, Wright, G.S.A, Antonyuk, S.V. | | Deposit date: | 2017-05-23 | | Release date: | 2018-01-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Study of the C-Terminal Domain of Nonstructural Protein 1 from Japanese Encephalitis Virus.
J. Virol., 92, 2018
|
|
6HNV
 
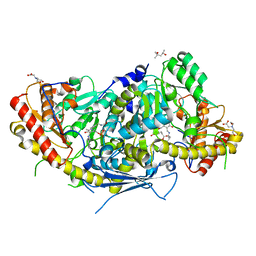 | | Crystal structure of aminotransferase Aro9 from C. Albicans with ligands | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINOHEXANEDIOIC ACID, 2-OXOADIPIC ACID, ... | | Authors: | Kiliszek, A, Rzad, K, Rypniewski, W, Milewski, S, Gabriel, I. | | Deposit date: | 2018-09-17 | | Release date: | 2019-02-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of aminotransferases Aro8 and Aro9 from Candida albicans and structural insights into their properties.
J.Struct.Biol., 205, 2019
|
|
6HVA
 
 | | Yeast 20S proteasome with human beta2i (1-53) in complex with 13 | | Descriptor: | (2~{S})-~{N}-[(2~{S})-1-[[(2~{S})-1-[4-(aminomethyl)phenyl]-4-methylsulfonyl-butan-2-yl]amino]-3-oxidanyl-1-oxidanylidene-propan-2-yl]-2-[[(2~{S})-2-azido-3-phenyl-propanoyl]amino]-4-methyl-pentanamide, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2018-10-10 | | Release date: | 2019-01-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure-Based Design of Inhibitors Selective for Human Proteasome beta 2c or beta 2i Subunits.
J.Med.Chem., 62, 2019
|
|
6HW9
 
 | | Yeast 20S proteasome in complex with 41b | | Descriptor: | (2~{S})-3-(4-methoxyphenyl)-~{N}-[(2~{S},3~{R})-4-methyl-1-(4-methylcyclohexyl)-3,4-bis(oxidanyl)pentan-2-yl]-2-[[(2~{S})-2-(2-morpholin-4-ylethanoylamino)propanoyl]amino]propanamide, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2018-10-11 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-Based Design of Inhibitors Selective for Human Proteasome beta 2c or beta 2i Subunits.
J.Med.Chem., 62, 2019
|
|
5J9R
 
 | |
