5G0T
 
 | | InhA in complex with a DNA encoded library hit | | Descriptor: | 1-benzyl-N-[cis-4-(2-{[(4-fluorophenyl)methyl][2-(methylamino)-2-oxoethyl]amino}-2-oxoethyl)cyclohexyl]-5-methyl-1H-1,2,3-triazole-4-carboxamide, ENOYL-[ACYL-CARRIER-PROTEIN] REDUCTASE [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Read, J.A, Breed, J. | | Deposit date: | 2016-03-22 | | Release date: | 2016-11-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Discovery of Cofactor-Specific, Bactericidal Mycobacterium Tuberculosis Inha Inhibitors Using DNA-Encoded Library Technology
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
6ZAV
 
 | | NO-bound copper nitrite reductase from Bradyrhizobium sp. ORS 375 (two-domain) at 1.19 A resolution (unrestrained, full matrix refinement by SHELX) | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, COPPER (II) ION, Copper-containing nitrite reductase, ... | | Authors: | Rose, S.L, Antonyuk, S.V, Sasaki, D, Yamashita, K, Hirata, K, Ueno, G, Ago, H, Eady, R.R, Tosha, T, Yamamoto, M, Hasnain, S.S. | | Deposit date: | 2020-06-05 | | Release date: | 2021-01-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | An unprecedented insight into the catalytic mechanism of copper nitrite reductase from atomic-resolution and damage-free structures.
Sci Adv, 7, 2021
|
|
9KTL
 
 | | Cryo-EM structure of reduced form of formate dehydrogenase from Rhodobacter aestuarii (RaFDH) with NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Zhang, K, Zhang, L. | | Deposit date: | 2024-12-02 | | Release date: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Mechanistic understanding on the catalytic preference of formate dehydrogenase from Rhodobacter aestuarii towards carbon dioxide reduction
To Be Published
|
|
6HEZ
 
 | | M tuberculosis DprE1 in complex with BTZ043 | | Descriptor: | 8-(hydroxyamino)-2-[(2S)-2-methyl-1,4-dioxa-8-azaspiro[4.5]dec-8-yl]-6-(trifluoromethyl)-4H-1,3-benzothiazin-4-one, Decaprenylphosphoryl-beta-D-ribose oxidase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Futterer, K, Batt, S.M, Besra, G.S. | | Deposit date: | 2018-08-21 | | Release date: | 2018-08-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Novel insight into the reaction of nitro, nitroso and hydroxylamino benzothiazinones and of benzoxacinones with Mycobacterium tuberculosis DprE1.
Sci Rep, 8, 2018
|
|
6HF3
 
 | | M tuberculosis DprE1 in complex with a covalently bound nitrobenzothiazinone | | Descriptor: | 2-(4-(cyclohexylmethyl)piperazin-1-yl)-8-nitro-6-(trifluoromethyl)-4H-benzo[e][1,3]thiazin-4-one, bound form, Decaprenylphosphoryl-beta-D-ribose oxidase, ... | | Authors: | Futterer, K, Batt, S.M, Besra, G.S. | | Deposit date: | 2018-08-21 | | Release date: | 2018-08-29 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Novel insight into the reaction of nitro, nitroso and hydroxylamino benzothiazinones and of benzoxacinones with Mycobacterium tuberculosis DprE1.
Sci Rep, 8, 2018
|
|
8B5Y
 
 | | SHP2 in complex with allosteric imidazopyrazine inhibitor | | Descriptor: | (1~{S})-1'-[5-[2-(trifluoromethyl)pyridin-3-yl]sulfanyl-3~{H}-imidazo[4,5-b]pyrazin-2-yl]spiro[1,3-dihydroindene-2,4'-piperidine]-1-amine, FORMIC ACID, Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Torrente, E, Petrocchi, A. | | Deposit date: | 2022-09-25 | | Release date: | 2023-01-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Discovery of a Novel Series of Imidazopyrazine Derivatives as Potent SHP2 Allosteric Inhibitors.
Acs Med.Chem.Lett., 14, 2023
|
|
6HOI
 
 | | Structure of Beclin1 LIR motif bound to GABARAPL1 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Beclin-1, ... | | Authors: | Mouilleron, S, Birgisdottir, A.B, Bhujbal, Z, Wirth, M, Sjottem, E, Evjen, G, Zhang, W, Lee, R, O'Reilly, N, Tooze, S, Lamark, T, Johansen, T. | | Deposit date: | 2018-09-17 | | Release date: | 2019-02-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Members of the autophagy class III phosphatidylinositol 3-kinase complex I interact with GABARAP and GABARAPL1 via LIR motifs.
Autophagy, 15, 2019
|
|
5F9N
 
 | |
9KSA
 
 | | Serine Beta-Lactamase OXA-48 in Complex with MBL/SBL Inhibitor CB1 | | Descriptor: | 2-oxidanyl-7-(pyrazol-1-ylmethyl)-3~{H}-1,4,2-benzodioxaborinine-8-carboxylic acid, Beta-lactamase | | Authors: | Li, G.-B, Wei, S.-Q. | | Deposit date: | 2024-11-29 | | Release date: | 2025-09-03 | | Method: | X-RAY DIFFRACTION (2.178 Å) | | Cite: | OXA-48 carbapenemase in complex with 7-((1H-pyrazol-1-yl)methyl)-2-hydroxy-2,3-dihydrobenzo[e][1,4,2]dioxaborinine-8-carboxylic acid
To Be Published
|
|
9COV
 
 | | Crystal structure of CYP3A4 bound to an inhibitor | | Descriptor: | Cytochrome P450 3A4, N-[(2S)-1-(1H-indol-3-yl)-3-{[(2R)-1-oxo-3-phenyl-1-{[3-(pyridin-3-yl)propyl]amino}propan-2-yl]sulfanyl}propan-2-yl]pyridine-3-carboxamide, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sevrioukova, I.F. | | Deposit date: | 2024-07-17 | | Release date: | 2025-01-08 | | Last modified: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Evaluation of Larger Side-Group Functionalities and the Side/End-Group Interplay in Ritonavir-Like Inhibitors of CYP3A4.
Chem.Biol.Drug Des., 105, 2025
|
|
9CU2
 
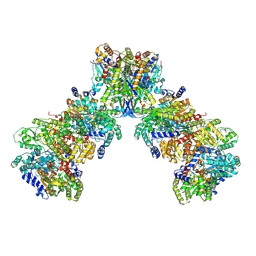 | | Azotobacter vinelandii filamentous 2:2:1 MoFeP:FeP:FeSII-Complex (C2 symmetry) | | Descriptor: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, ADENOSINE-5'-DIPHOSPHATE, FE (III) ION, ... | | Authors: | Narehood, S.M, Cook, B.D, Srisantitham, S, Eng, V.H, Shiau, A, Britt, R.D, Herzik, M.A, Tezcan, F.A. | | Deposit date: | 2024-07-25 | | Release date: | 2025-01-15 | | Last modified: | 2025-08-20 | | Method: | ELECTRON MICROSCOPY (2.27 Å) | | Cite: | Structural basis for the conformational protection of nitrogenase from O 2 .
Nature, 637, 2025
|
|
5JE6
 
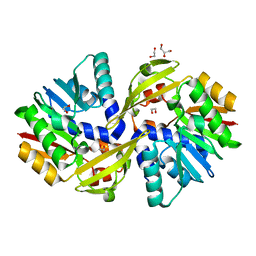 | | Crystal structure of Burkholderia glumae ToxA | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Methyl transferase | | Authors: | Fenwick, M.K, Philmus, B, Begley, T.P, Ealick, S.E. | | Deposit date: | 2016-04-17 | | Release date: | 2016-05-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.571 Å) | | Cite: | Burkholderia glumae ToxA Is a Dual-Specificity Methyltransferase That Catalyzes the Last Two Steps of Toxoflavin Biosynthesis.
Biochemistry, 55, 2016
|
|
9CU1
 
 | | Azotobacter vinelandii filamentous 2:2:1 MoFeP:FeP:FeSII-Complex (termini; C1 symmetry) | | Descriptor: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, ADENOSINE-5'-DIPHOSPHATE, FE (III) ION, ... | | Authors: | Narehood, S.M, Cook, B.D, Srisantitham, S, Eng, V.H, Shiau, A, Britt, R.D, Herzik, M.A, Tezcan, F.A. | | Deposit date: | 2024-07-25 | | Release date: | 2025-01-15 | | Last modified: | 2025-08-20 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Structural basis for the conformational protection of nitrogenase from O 2 .
Nature, 637, 2025
|
|
5JGU
 
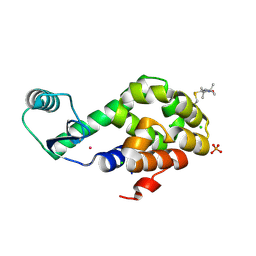 | | Spin-Labeled T4 Lysozyme Construct R119V1 | | Descriptor: | CHLORIDE ION, Endolysin, PHOSPHATE ION, ... | | Authors: | Balo, A.R, Feyrer, H, Ernst, O.P. | | Deposit date: | 2016-04-20 | | Release date: | 2017-02-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.468 Å) | | Cite: | Toward Precise Interpretation of DEER-Based Distance Distributions: Insights from Structural Characterization of V1 Spin-Labeled Side Chains.
Biochemistry, 55, 2016
|
|
5JI8
 
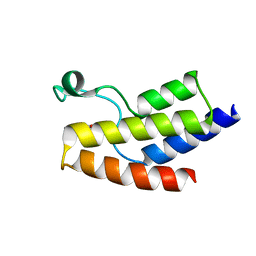 | | Crystal structure of the BRD9 bromodomain and hit 1 | | Descriptor: | 2-amino-1,3-benzothiazole-6-carboxamide, Bromodomain-containing protein 9 | | Authors: | Wang, N, Li, F, Bao, H, Li, J, Wu, J, Ruan, K. | | Deposit date: | 2016-04-22 | | Release date: | 2016-06-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | NMR Fragment Screening Hit Induces Plasticity of BRD7/9 Bromodomains
Chembiochem, 17, 2016
|
|
6YJ4
 
 | | Structure of Yarrowia lipolytica complex I at 2.7 A | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, Acyl carrier protein ACPM1 of NADH:Ubiquinone Oxidoreductase (Complex I), Acyl carrier protein ACPM2 of NADH:Ubiquinone Oxidoreductase (Complex I), ... | | Authors: | Hirst, J, Grba, D. | | Deposit date: | 2020-04-02 | | Release date: | 2020-08-12 | | Last modified: | 2025-10-01 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Mitochondrial complex I structure reveals ordered water molecules for catalysis and proton translocation.
Nat.Struct.Mol.Biol., 27, 2020
|
|
5IZV
 
 | | Crystal structure of the legionella pneumophila effector protein RavZ - F222 | | Descriptor: | Uncharacterized protein RavZ | | Authors: | Kwon, D.H, Kim, L, Kim, B.-W, Hong, S.B, Song, H.K. | | Deposit date: | 2016-03-26 | | Release date: | 2016-11-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.814 Å) | | Cite: | The 1:2 complex between RavZ and LC3 reveals a mechanism for deconjugation of LC3 on the phagophore membrane
Autophagy, 13, 2017
|
|
6FBK
 
 | | Crystal structure of the human WNK2 CCT-like 1 domain in complex with a WNK1 RFXV peptide | | Descriptor: | Serine/threonine-protein kinase WNK1, Serine/threonine-protein kinase WNK2 | | Authors: | Pinkas, D.M, Bufton, J.C, Burgess-Brown, N.A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A.N. | | Deposit date: | 2017-12-19 | | Release date: | 2018-01-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.743 Å) | | Cite: | Crystal structure of the human WNK2 CCT-like 1 domain in complex with a WNK1 RFXV peptide
To Be Published
|
|
3LPL
 
 | | E. coli pyruvate dehydrogenase complex E1 component E571A mutant | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Furey, W. | | Deposit date: | 2010-02-05 | | Release date: | 2010-03-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Communication between thiamin cofactors in the Escherichia coli pyruvate dehydrogenase complex E1 component active centers: evidence for a "direct pathway" between the 4'-aminopyrimidine N1' atoms.
J.Biol.Chem., 285, 2010
|
|
5CXD
 
 | | 1.75 Angstrom resolution crystal structure of the apo-form acyl-carrier-protein synthase (AcpS) (acpS; purification tag off) from Staphylococcus aureus subsp. aureus COL in the I4 space group | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, Holo-[acyl-carrier-protein] synthase, ... | | Authors: | Halavaty, A.S, Minasov, G, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-07-28 | | Release date: | 2015-08-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | 1.75 Angstrom resolution crystal structure of the apo-form acyl-carrier-protein synthase (AcpS) (acpS; purification tag off) from Staphylococcus aureus subsp. aureus COL in the I4 space group
To Be Published
|
|
9CUM
 
 | | Q67H mutant of R67 DHFR complexed with Congo Red | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, Dihydrofolate reductase type 2, ... | | Authors: | Narayana, N, Narendra, A.N. | | Deposit date: | 2024-07-26 | | Release date: | 2025-02-19 | | Last modified: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Crystal structure of the plasmid-encoded R67 dihydrofolate reductase complexed with Congo red an amyloid binding dye.
Sci Rep, 15, 2025
|
|
7XZG
 
 | | High resolution crystal Structure of Glyceraldehyde-3-Phosphate Dehydrogenase from Candida albicans complexed with NAD+ | | Descriptor: | 1,2-ETHANEDIOL, Glyceraldehyde-3-phosphate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Cao, H, Meng, J, Ren, Y, Wan, J. | | Deposit date: | 2022-06-02 | | Release date: | 2023-06-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.879 Å) | | Cite: | High resolution crystal Structure of Glyceraldehyde-3-Phosphate Dehydrogenase from Candida albicans complexed with NAD+
To Be Published
|
|
7RGJ
 
 | | DfrA1 complexed with NADPH and 5-(3-(7-(4-(aminomethyl)phenyl)benzo[d][1,3]dioxol-5-yl)but-1-yn-1-yl)-6-ethylpyrimidine-2,4-diamine (UCP1223) | | Descriptor: | 5-[(3R)-3-{7-[4-(aminomethyl)phenyl]-2H-1,3-benzodioxol-5-yl}but-1-yn-1-yl]-6-ethylpyrimidine-2,4-diamine, 5-[(3S)-3-{7-[4-(aminomethyl)phenyl]-2H-1,3-benzodioxol-5-yl}but-1-yn-1-yl]-6-ethylpyrimidine-2,4-diamine, CALCIUM ION, ... | | Authors: | Lombardo, M.N, Wright, D.L. | | Deposit date: | 2021-07-15 | | Release date: | 2022-05-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Structure-guided functional studies of plasmid-encoded dihydrofolate reductases reveal a common mechanism of trimethoprim resistance in Gram-negative pathogens.
Commun Biol, 5, 2022
|
|
3LQ2
 
 | | E. coli pyruvate dehydrogenase complex E1 E235A mutant with low TDP concentration | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Furey, W. | | Deposit date: | 2010-02-08 | | Release date: | 2010-03-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Communication between thiamin cofactors in the Escherichia coli pyruvate dehydrogenase complex E1 component active centers: evidence for a "direct pathway" between the 4'-aminopyrimidine N1' atoms.
J.Biol.Chem., 285, 2010
|
|
5HYP
 
 | |
