2O21
 
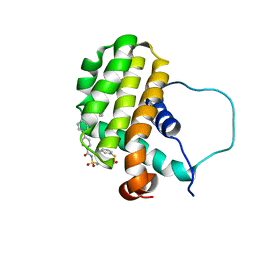 | | Solution structure of the anti-apoptotic protein Bcl-2 in complex with an acyl-sulfonamide-based ligand | | Descriptor: | 3-NITRO-N-{4-[2-(2-PHENYLETHYL)-1,3-BENZOTHIAZOL-5-YL]BENZOYL}-4-{[2-(PHENYLSULFANYL)ETHYL]AMINO}BENZENESULFONAMIDE, Apoptosis regulator Bcl-2 | | Authors: | Bruncko, M, Oost, T.K, Belli, B.A, Ding, H, Joseph, M.K, Kunzer, A, Martineau, D, McClellan, W.J, Mitten, M, Ng, S.C, Nimmer, P.M, Oltersdorf, T, Park, C.M, Petros, A.M, Shoemaker, A.R, Song, X, Wang, X, Wendt, M.D, Zhang, H, Fesik, S.W, Rosenberg, S.H, Elmore, S.W. | | Deposit date: | 2006-11-29 | | Release date: | 2007-02-27 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Studies Leading to Potent, Dual Inhibitors of Bcl-2 and Bcl-xL.
J.Med.Chem., 50, 2007
|
|
3D1P
 
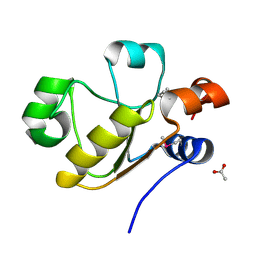 | | Atomic resolution structure of uncharacterized protein from Saccharomyces cerevisiae | | Descriptor: | ACETATE ION, CHLORIDE ION, Putative thiosulfate sulfurtransferase YOR285W | | Authors: | Nocek, B, Evdokimova, E, Kudritska, M, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-05-06 | | Release date: | 2008-07-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | Atomic resolution structure of uncharacterized protein from Saccharomyces cerevisiae.
To be Published
|
|
6W4T
 
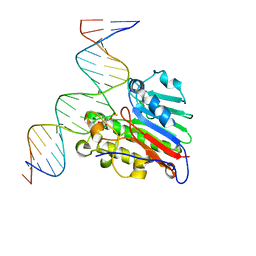 | | APE1 Y269A phosphorothioate substrate complex with abasic DNA | | Descriptor: | DNA (5'-D(*GP*GP*AP*TP*CP*CP*GP*TP*CP*GP*GP*AP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), DNA (5'-D(P*GP*CP*TP*GP*AP*TP*GP*CP*GP*TP*(48Z)P*CP*GP*AP*CP*GP*GP*AP*TP*CP*C)-3'), DNA-(apurinic or apyrimidinic site) lyase | | Authors: | Freudenthal, B.D, Hoitsma, N.M. | | Deposit date: | 2020-03-11 | | Release date: | 2020-06-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | AP-endonuclease 1 sculpts DNA through an anchoring tyrosine residue on the DNA intercalating loop.
Nucleic Acids Res., 48, 2020
|
|
4TZ3
 
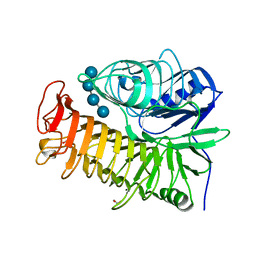 | | Ensemble refinement of the E502A variant of sacteLam55A from Streptomyces sp. SirexAA-E in complex with laminaritetraose | | Descriptor: | 1,2-ETHANEDIOL, Putative secreted protein, beta-D-glucopyranose, ... | | Authors: | Bianchetti, C.M, Takasuka, T.E, Yik, E.J, Bergeman, L.F, Fox, B.G. | | Deposit date: | 2014-07-09 | | Release date: | 2015-03-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Active site and laminarin binding in glycoside hydrolase family 55.
J.Biol.Chem., 290, 2015
|
|
3D2Y
 
 | | Complex of the N-acetylmuramyl-L-alanine amidase AmiD from E.coli with the substrate anhydro-N-acetylmuramic acid-L-Ala-D-gamma-Glu-L-Lys | | Descriptor: | Anhydro-N-acetylmuramic acid-L-Ala-D-gamma-Glu-L-Lys, GLYCEROL, N-acetylmuramoyl-L-alanine amidase amiD | | Authors: | Kerff, F, Petrella, S, Herman, R, Sauvage, E, Mercier, F, Luxen, A, Frere, J.M, Joris, B, Charlier, P. | | Deposit date: | 2008-05-09 | | Release date: | 2009-06-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Specific Structural Features of the N-Acetylmuramoyl-l-Alanine Amidase AmiD from Escherichia coli and Mechanistic Implications for Enzymes of This Family.
J.Mol.Biol., 397, 2010
|
|
1T6L
 
 | | Crystal Structure of the Human Cytomegalovirus DNA Polymerase Subunit, UL44 | | Descriptor: | DNA polymerase processivity factor | | Authors: | Appleton, B.A, Loregian, A, Filman, D.J, Coen, D.M, Hogle, J.M. | | Deposit date: | 2004-05-06 | | Release date: | 2004-08-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The Cytomegalovirus DNA Polymerase Subunit UL44 Forms a C Clamp-Shaped Dimer.
Mol.Cell, 15, 2004
|
|
4KEC
 
 | | SbHCT-complex form | | Descriptor: | (3R,4S,5R)-3,4-dihydroxy-5-{[(2E)-3-(4-hydroxyphenyl)prop-2-enoyl]oxy}cyclohex-1-ene-1-carboxylic acid, COENZYME A, Hydroxycinnamoyl-CoA:shikimate hydroxycinnamoyl transferase | | Authors: | Walker, A.M, Hayes, R.P, Youn, B, Vermerris, W, Sattler, S.E, Kang, C. | | Deposit date: | 2013-04-25 | | Release date: | 2013-05-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Elucidation of the structure and reaction mechanism of sorghum hydroxycinnamoyltransferase and its structural relationship to other coenzyme a-dependent transferases and synthases.
Plant Physiol., 162, 2013
|
|
1TJ0
 
 | | Crystal structure of E. coli PutA proline dehydrogenase domain (residues 86-669) co-crystallized with L-lactate | | Descriptor: | (2S)-2-HYDROXYPROPANOIC ACID, Bifunctional putA protein, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Tanner, J.J, Zhang, M, White, T.A, Schuermann, J.P, Baban, B.A, Becker, D.F. | | Deposit date: | 2004-06-02 | | Release date: | 2004-10-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of the Escherichia coli PutA proline dehydrogenase domain in complex with competitive inhibitors
Biochemistry, 43, 2004
|
|
7FHE
 
 | | Structure of prenyltransferase mutant Q295F from Streptomyces sp. (strain CL190) | | Descriptor: | Prenyltransferase | | Authors: | Xue, B, Lim, K.J.H, Hartono, Y.D, Go, M.D.K, Fan, H, Yew, W.S. | | Deposit date: | 2021-07-29 | | Release date: | 2022-06-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-Guided Engineering of Prenyltransferase NphB for High-Yield and Regioselective Cannabinoid Production.
Acs Catalysis, 12, 2022
|
|
7FHC
 
 | | Structure of prenyltransferase mutant V49W from Streptomyces sp. (strain CL190) | | Descriptor: | Prenyltransferase | | Authors: | Xue, B, Lim, K.J.H, Hartono, Y.D, Go, M.D.K, Fan, H, Yew, W.S. | | Deposit date: | 2021-07-29 | | Release date: | 2022-06-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structure-Guided Engineering of Prenyltransferase NphB for High-Yield and Regioselective Cannabinoid Production.
Acs Catalysis, 12, 2022
|
|
7FHB
 
 | | Structure of prenyltransferase from Streptomyces sp. (strain CL190) with bound GPP | | Descriptor: | GERANYL DIPHOSPHATE, Prenyltransferase | | Authors: | Xue, B, Lim, K.J.H, Hartono, Y.D, Go, M.D.K, Fan, H, Yew, W.S. | | Deposit date: | 2021-07-29 | | Release date: | 2022-06-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-Guided Engineering of Prenyltransferase NphB for High-Yield and Regioselective Cannabinoid Production.
Acs Catalysis, 12, 2022
|
|
1TK5
 
 | | T7 DNA polymerase binary complex with 8 oxo guanosine in the templating strand | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5'-D(*CP*CP*CP*(8OG)P*CP*TP*GP*GP*CP*AP*CP*TP*GP*GP*CP*CP*GP*TP*CP*GP*TP*TP*TP*TP*CP*G)-3', 5'-D(*CP*GP*AP*AP*A*GP*CP*CP*AP*GP*TP*GP*CP*CP*AP*(DDG)P*TP*GP*CP*AP*A)-3', ... | | Authors: | Brieba, L.G, Eichman, B.F, Kokoska, R.J, Doublie, S, Kunkel, T.A, Ellenberger, T. | | Deposit date: | 2004-06-08 | | Release date: | 2004-08-31 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for the dual coding potential of 8-oxoguanosine by a high-fidelity DNA polymerase.
Embo J., 23, 2004
|
|
4KEK
 
 | |
5B5U
 
 | | Crystal structure of truncated Pyrococcus furiosus L-asparaginase with peptide | | Descriptor: | ASPARTIC ACID, CHLORIDE ION, GLYCEROL, ... | | Authors: | Sharma, P, Tomar, R, Nath, S.K, Kundu, B, Ashish, F. | | Deposit date: | 2016-05-23 | | Release date: | 2017-05-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Heat induces end to end repetitive association in P. furiosus L-asparaginase which enables its thermophilic property.
Sci Rep, 10, 2020
|
|
1T79
 
 | | Crystal structure of the androgen receptor ligand binding domain in complex with a FxxLW motif | | Descriptor: | 5-ALPHA-DIHYDROTESTOSTERONE, Androgen receptor, FxxLW motif peptide | | Authors: | Hur, E, Pfaff, S.J, Payne, E.S, Gron, H, Buehrer, B.M, Fletterick, R.J. | | Deposit date: | 2004-05-08 | | Release date: | 2004-08-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Recognition and accommodation at the androgen receptor coactivator binding interface.
Plos Biol., 2, 2004
|
|
3D49
 
 | | Thrombin Inhibition | | Descriptor: | BENZAMIDINE, Hirudin variant-1, SODIUM ION, ... | | Authors: | Baum, B, Heine, A, Klebe, G. | | Deposit date: | 2008-05-14 | | Release date: | 2009-04-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Understanding Thrombin Inhibition
To be Published
|
|
1T21
 
 | | Structural basis for degenerate recognition of HIV peptide variants by cytotoxic lymphocyte, variant SL9, monoclinic crystal | | Descriptor: | Beta-2-microglobulin, GAG PEPTIDE, HLA class I histocompatibility antigen, ... | | Authors: | Martinez-Hackert, E, Anikeeva, N, Kalams, S.A, Walker, B.D, Hendrickson, W.A, Sykulev, Y. | | Deposit date: | 2004-04-19 | | Release date: | 2005-09-06 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structural Basis for Degenerate Recognition of Natural HIV Peptide Variants by Cytotoxic Lymphocytes.
J.Biol.Chem., 281, 2006
|
|
7F5D
 
 | | Crystal structure of BPTF-BRD with ligand DC-BPi-03 bound | | Descriptor: | 6-(1H-indol-5-yl)-N-methyl-2-methylsulfonyl-pyrimidin-4-amine, Nucleosome-remodeling factor subunit BPTF | | Authors: | Lu, T, Lu, H.B, Wang, J, Lin, H, Lu, W, Luo, C. | | Deposit date: | 2021-06-21 | | Release date: | 2022-06-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.57150865 Å) | | Cite: | Discovery and Optimization of Small-Molecule Inhibitors for the BPTF Bromodomains Proteins
To Be Published
|
|
8PZK
 
 | | Crystal structure of the Orange Carotenoid Protein 2 (OCP2) from Gloeocapsa sp. PCC 7428 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, AZIDE ION, GLYCEROL, ... | | Authors: | Boyko, K.M, Varfolomeeva, L.A, Slonimskiy, Y.B, Maksimov, E.G, Popov, V.O, Sluchanko, N.N. | | Deposit date: | 2023-07-27 | | Release date: | 2024-01-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural framework for the understanding spectroscopic and functional signatures of the cyanobacterial Orange Carotenoid Protein families.
Int.J.Biol.Macromol., 254, 2024
|
|
4KLF
 
 | | DNA polymerase beta matched reactant complex with Mg2+, 20 s | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, 5'-D(*CP*CP*GP*AP*CP*GP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3', 5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*CP*C)-3', ... | | Authors: | Freudenthal, B.D, Beard, W.A, Shock, D.D, Wilson, S.H. | | Deposit date: | 2013-05-07 | | Release date: | 2013-07-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Observing a DNA polymerase choose right from wrong.
Cell(Cambridge,Mass.), 154, 2013
|
|
1T45
 
 | | STRUCTURAL BASIS FOR THE AUTOINHIBITION AND STI-571 INHIBITION OF C-KIT TYROSINE KINASE | | Descriptor: | Homo sapiens v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog | | Authors: | Mol, C.D, Dougan, D.R, Schneider, T.R, Skene, R.J, Kraus, M.L, Scheibe, D.N, Snell, G.P, Zou, H, Sang, B.C, Wilson, K.P. | | Deposit date: | 2004-04-28 | | Release date: | 2004-06-15 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for the autoinhibition and STI-571 inhibition of c-Kit tyrosine kinase.
J.Biol.Chem., 279, 2004
|
|
7FHF
 
 | | Structure of prenyltransferase mutant V49W/Y288F/Q295F from Streptomyces sp. (strain CL190) | | Descriptor: | Prenyltransferase | | Authors: | Xue, B, Lim, K.J.H, Hartono, Y.D, Go, M.D.K, Fan, H, Yew, W.S. | | Deposit date: | 2021-07-29 | | Release date: | 2022-06-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-Guided Engineering of Prenyltransferase NphB for High-Yield and Regioselective Cannabinoid Production.
Acs Catalysis, 12, 2022
|
|
2OCB
 
 | | Crystal structure of human RAB9B in complex with a GTP analogue | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Ras-related protein Rab-9B, ... | | Authors: | Hong, B, Shen, L, Walker, J.R, Tempel, W, Landry, R, Arrowsmith, C.H, Edwards, A.M, Sundstrom, M, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-12-20 | | Release date: | 2007-01-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of human RAB9B in complex with a GTP analogue
To be Published
|
|
3CSP
 
 | | Crystal structure of the DM2 mutant of myelin oligodendrocyte glycoprotein | | Descriptor: | Myelin-oligodendrocyte glycoprotein | | Authors: | Breithaupt, C, Schafer, B, Pellkofer, H, Huber, R, Linington, C, Jacob, U. | | Deposit date: | 2008-04-10 | | Release date: | 2008-10-21 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Demyelinating myelin oligodendrocyte glycoprotein-specific autoantibody response is focused on one dominant conformational epitope region in rodents
J.Immunol., 181, 2008
|
|
6ZLM
 
 | | Dihydrolipoyllysine-residue acetyltransferase component of fungal pyruvate dehydrogenase complex with protein X bound | | Descriptor: | Dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase complex, mitochondrial, Pyruvate dehydrogenase X component | | Authors: | Forsberg, B.O, Aibara, S, Howard, R.J, Mortezaei, N, Lindahl, E. | | Deposit date: | 2020-06-30 | | Release date: | 2020-09-23 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Arrangement and symmetry of the fungal E3BP-containing core of the pyruvate dehydrogenase complex.
Nat Commun, 11, 2020
|
|
