6H6L
 
 | |
5L05
 
 | |
9LIG
 
 | | Cryo-EM structure of Saccharomyces cerevisiae Mitochondrial Respiratory Complex II in Y19315-bound state | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 3-[bis(fluoranyl)methyl]-~{N}-cyclopropyl-~{N}-[2-(2,4-dichlorophenyl)prop-2-enyl]-5-fluoranyl-1-methyl-pyrazole-4-carboxamide, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Li, Z.W, Ye, Y, Yang, G.F. | | Deposit date: | 2025-01-14 | | Release date: | 2025-09-03 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Cryo-EM structure of Saccharomyces cerevisiae Mitochondrial Respiratory Complex II in Y19315-bound state
To Be Published
|
|
9KUW
 
 | | Cryo-EM structure of dimeric APJR and two Beta-arrestins complex with small molecules | | Descriptor: | (1~{S},2~{S})-~{N}-[4-(2,6-dimethoxyphenyl)-5-(6-methylpyridin-2-yl)-1,2,4-triazol-3-yl]-1-(5-methylpyrimidin-2-yl)-1-oxidanyl-propane-2-sulfonamide, Apelin receptor, Beta arrestion-scFv30, ... | | Authors: | Yue, Y, Wu, L.J, Xu, F. | | Deposit date: | 2024-12-04 | | Release date: | 2025-09-17 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | Mechanistic insights into the versatile stoichiometry and biased signaling of the apelin receptor-arrestin complex.
Nat Commun, 16, 2025
|
|
5A7S
 
 | | Crystal structure of human JMJD2A in complex with compound 44 | | Descriptor: | 1,2-ETHANEDIOL, 2-(5-acetamido-2-oxidanyl-phenyl)pyridine-4-carboxylic acid, DIMETHYL SULFOXIDE, ... | | Authors: | Nowak, R, Velupillai, S, Krojer, T, Gileadi, C, Johansson, C, Korczynska, M, Le, D.D, Younger, N, Gregori-Puigjane, E, Tumber, A, Iwasa, E, Pollock, S.B, Ortiz Torres, I, Kopec, J, Tallant, C, Froese, S, von Delft, F, Arrowsmith, C.H, Bountra, C, Edwards, A, Shoichet, B.K, Fujimori, D.G, Oppermann, U. | | Deposit date: | 2015-07-09 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Docking and Linking of Fragments to Discover Jumonji Histone Demethylase Inhibitors.
J.Med.Chem., 59, 2016
|
|
9KUV
 
 | | Mechanistic insights into the versatile stoichiometry and biased signaling of the apelin receptor-arrestin complex | | Descriptor: | (1~{S},2~{S})-~{N}-[4-(2,6-dimethoxyphenyl)-5-(6-methylpyridin-2-yl)-1,2,4-triazol-3-yl]-1-(5-methylpyrimidin-2-yl)-1-oxidanyl-propane-2-sulfonamide, Apelin Receptor, Beta-arrestin-scFv30, ... | | Authors: | Yue, Y, Wu, L.J, Xu, F. | | Deposit date: | 2024-12-04 | | Release date: | 2025-09-17 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Mechanistic insights into the versatile stoichiometry and biased signaling of the apelin receptor-arrestin complex.
Nat Commun, 16, 2025
|
|
9KUX
 
 | | Cryo-EM structure of dimeric APJR and one Beta-arrestin complex with small molecules | | Descriptor: | (1~{S},2~{S})-~{N}-[4-(2,6-dimethoxyphenyl)-5-(6-methylpyridin-2-yl)-1,2,4-triazol-3-yl]-1-(5-methylpyrimidin-2-yl)-1-oxidanyl-propane-2-sulfonamide, Apelin receptor, Beta arrestin-scFv30 | | Authors: | Yue, Y, Wu, L.J, Xu, F. | | Deposit date: | 2024-12-04 | | Release date: | 2025-09-17 | | Method: | ELECTRON MICROSCOPY (3.57 Å) | | Cite: | Mechanistic insights into the versatile stoichiometry and biased signaling of the apelin receptor-arrestin complex.
Nat Commun, 16, 2025
|
|
9FU3
 
 | | Closed CODH/ACS in the methylated state | | Descriptor: | CO-methylating acetyl-CoA synthase, Carbon monoxide dehydrogenase, Fe(3)-Ni(1)-S(4) cluster, ... | | Authors: | Ruickoldt, J, Wendler, P, Dobbek, H. | | Deposit date: | 2024-06-26 | | Release date: | 2025-07-09 | | Last modified: | 2025-09-17 | | Method: | ELECTRON MICROSCOPY (2.35 Å) | | Cite: | Ligand binding to a Ni-Fe cluster orchestrates conformational changes of the CO-dehydrogenase-acetyl-CoA synthase complex.
Nat Catal, 8, 2025
|
|
6HU3
 
 | | Crystal structure of Schistosoma mansoni HDAC8 complexed with a triazole hydroxamate inhibitor | | Descriptor: | 1-[5-chloranyl-2-(4-fluoranylphenoxy)phenyl]-~{N}-oxidanyl-1,2,3-triazole-4-carboxamide, DIMETHYLFORMAMIDE, GLYCEROL, ... | | Authors: | Shaik, T.B, Marek, M, Romier, C. | | Deposit date: | 2018-10-05 | | Release date: | 2018-10-31 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.655 Å) | | Cite: | Characterization of Histone Deacetylase 8 (HDAC8) Selective Inhibition Reveals Specific Active Site Structural and Functional Determinants.
J. Med. Chem., 61, 2018
|
|
6WUY
 
 | | Crystal Structure of Recombinant Human Acetylcholinesterase In Complex with GA and HI-6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | McGuire, J.R, Bester, S.M, Pegan, S.D, Height, J.J. | | Deposit date: | 2020-05-05 | | Release date: | 2021-02-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Structural and Biochemical Insights into the Inhibition of Human Acetylcholinesterase by G-Series Nerve Agents and Subsequent Reactivation by HI-6.
Chem.Res.Toxicol., 34, 2021
|
|
7RGK
 
 | | DfrA5 complexed with NADPH and 5-(3-(7-(4-(aminomethyl)phenyl)benzo[d][1,3]dioxol-5-yl)but-1-yn-1-yl)-6-ethylpyrimidine-2,4-diamine (UCP1223) | | Descriptor: | 5-[(3R)-3-{7-[4-(aminomethyl)phenyl]-2H-1,3-benzodioxol-5-yl}but-1-yn-1-yl]-6-ethylpyrimidine-2,4-diamine, Dihydrofolate reductase type 5, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Lombardo, M.N, Wright, D.L. | | Deposit date: | 2021-07-15 | | Release date: | 2022-05-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structure-guided functional studies of plasmid-encoded dihydrofolate reductases reveal a common mechanism of trimethoprim resistance in Gram-negative pathogens.
Commun Biol, 5, 2022
|
|
7RAD
 
 | | Crystal Structure Analysis of ALDH1B1 | | Descriptor: | 1,2-ETHANEDIOL, 3-(2-methoxyphenyl)-1-(4-phenylphenyl)-6,7,8,9-tetrahydro-5~{H}-imidazo[1,2-a][1,3]diazepine, Aldehyde dehydrogenase X, ... | | Authors: | Fernandez, D, Chen, J.K. | | Deposit date: | 2021-07-01 | | Release date: | 2022-06-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Inhibitors targeted to aldehyde dehydrogenase
Nat.Chem.Biol., 2022
|
|
5NQD
 
 | | Arsenite oxidase AioAB from Rhizobium sp. str. NT-26 mutant AioBF108A | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, ... | | Authors: | Santos-Silva, T, Romao, M, Vieira, M, Marques, A.T. | | Deposit date: | 2017-04-20 | | Release date: | 2018-05-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Arsenite oxidase AioAB from Rhizobium sp. str. NT-26 mutant AioBF108A
To Be Published
|
|
6QZH
 
 | | Structure of the human CC Chemokine Receptor 7 in complex with the intracellular allosteric antagonist Cmp2105 and the insertion protein Sialidase NanA | | Descriptor: | 3-[[4-[[(1~{R})-2,2-dimethyl-1-(5-methylfuran-2-yl)propyl]amino]-1,1-bis(oxidanylidene)-1,2,5-thiadiazol-3-yl]amino]-~{N},~{N},6-trimethyl-2-oxidanyl-benzamide, C-C chemokine receptor type 7,Sialidase A,C-C chemokine receptor type 7, D(-)-TARTARIC ACID, ... | | Authors: | Jaeger, K, Bruenle, S, Weinert, T, Guba, W, Muehle, J, Miyazaki, T, Weber, M, Furrer, A, Haenggi, N, Tetaz, T, Huang, C.Y, Mattle, D, Vonach, J.M, Gast, A, Kuglstatter, A, Rudolph, M.G, Nogly, P, Benz, J, Dawson, R.J.P, Standfuss, J. | | Deposit date: | 2019-03-11 | | Release date: | 2019-09-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis for Allosteric Ligand Recognition in the Human CC Chemokine Receptor 7.
Cell, 178, 2019
|
|
5N2S
 
 | | Crystal structure of stabilized A1 receptor in complex with PSB36 at 3.3A resolution | | Descriptor: | 1-butyl-3-(3-oxidanylpropyl)-8-[(1~{R},5~{S})-3-tricyclo[3.3.1.0^{3,7}]nonanyl]-7~{H}-purine-2,6-dione, SULFATE ION, Soluble cytochrome b562,Adenosine receptor A1 | | Authors: | Cheng, R.K.Y, Segala, E, Robertson, N, Deflorian, F, Dore, A.S, Errey, J.C, Fiez-Vandal, C, Marshall, F.H, Cooke, R.M. | | Deposit date: | 2017-02-08 | | Release date: | 2017-07-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.303 Å) | | Cite: | Structures of Human A1 and A2A Adenosine Receptors with Xanthines Reveal Determinants of Selectivity.
Structure, 25, 2017
|
|
9GR8
 
 | | Crystal structure of the N-terminal kinase domain from Saccharomyces cerevisiae Vip1 in complex with ADP. | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, Inositol hexakisphosphate and diphosphoinositol-pentakisphosphate kinase | | Authors: | Lee, K, Raia, P, Hothorn, M. | | Deposit date: | 2024-09-11 | | Release date: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | A small signaling domain controls PPIP5K phosphatase activity in phosphate homeostasis.
Nat Commun, 16, 2025
|
|
8ZI5
 
 | |
7REB
 
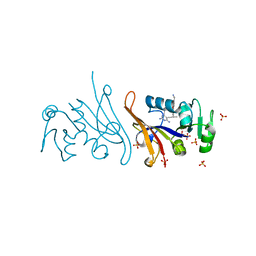 | | E. coli dihydrofolate reductase complexed with 5-(3-(7-(4-(aminomethyl)phenyl)benzo[d][1,3]dioxol-5-yl)but-1-yn-1-yl)-6-ethylpyrimidine-2,4-diamine (UCP1223) | | Descriptor: | 5-[(3R)-3-{7-[4-(aminomethyl)phenyl]-2H-1,3-benzodioxol-5-yl}but-1-yn-1-yl]-6-ethylpyrimidine-2,4-diamine, Dihydrofolate reductase, SULFATE ION | | Authors: | Lombardo, M.N, Wright, D.L. | | Deposit date: | 2021-07-12 | | Release date: | 2022-07-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structure-guided functional studies of plasmid-encoded dihydrofolate reductases reveal a common mechanism of trimethoprim resistance in Gram-negative pathogens.
Commun Biol, 5, 2022
|
|
6R4Y
 
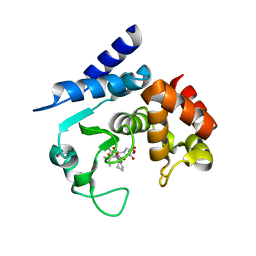 | |
8OON
 
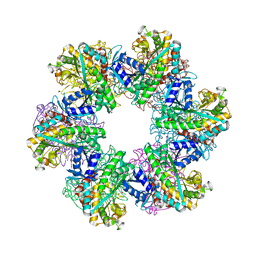 | | Glutamine synthetase from Methanothermococcus thermolithotrophicus at a resolution of 2.43 A | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Glutamine synthetase from Methanothermococcus thermolithotrophicus, ... | | Authors: | Mueller, M.-C, Lemaire, O.N, Wagner, T. | | Deposit date: | 2023-04-05 | | Release date: | 2024-01-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Differences in regulation mechanisms of glutamine synthetases from methanogenic archaea unveiled by structural investigations.
Commun Biol, 7, 2024
|
|
5XH6
 
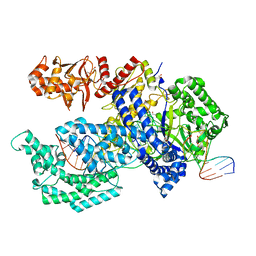 | | Crystal structure of the Acidaminococcus sp. BV3L6 Cpf1 RVR variant in complex with crRNA and target DNA (TATA PAM) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, CRISPR-associated endonuclease Cpf1, ... | | Authors: | Nishimasu, H, Yamano, T, Ishitani, R, Nureki, O. | | Deposit date: | 2017-04-19 | | Release date: | 2017-06-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for the Altered PAM Recognition by Engineered CRISPR-Cpf1
Mol. Cell, 67, 2017
|
|
7RQ0
 
 | |
7UWT
 
 | | Structure of Oxygen-Insensitive NAD(P)H-dependent Nitroreductase NfsB_Vv F70A/F108Y (NTR 2.0) in complex with FMN at 1.85 Angstroms resolution | | Descriptor: | ACETATE ION, Dihydropteridine reductase, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Sharrock, A.V, Arcus, V, Mumm, J.S, Ackerley, D.F. | | Deposit date: | 2022-05-03 | | Release date: | 2022-05-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The Crystal Structure of Engineered Nitroreductase NTR 2.0 and Impact of F70A and F108Y Substitutions on Substrate Specificity.
Int J Mol Sci, 24, 2023
|
|
5UBB
 
 | | Crystal structure of human alpha N-terminal protein methyltransferase 1B | | Descriptor: | Alpha N-terminal protein methyltransferase 1B, S-ADENOSYLMETHIONINE, UNKNOWN ATOM OR ION | | Authors: | Dong, C, Zhu, L, Tempel, W, Dong, A, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-12-20 | | Release date: | 2017-03-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | An asparagine/glycine switch governs product specificity of human N-terminal methyltransferase NTMT2.
Commun Biol, 1, 2018
|
|
5TZP
 
 | | Crystal structure of FPV039:Bik BH3 complex | | Descriptor: | 1,2-ETHANEDIOL, Bcl-2-like protein FPV039, Bik, ... | | Authors: | Anasir, M.I, Kvansakul, M. | | Deposit date: | 2016-11-22 | | Release date: | 2017-05-03 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural basis of apoptosis inhibition by the fowlpox virus protein FPV039.
J. Biol. Chem., 292, 2017
|
|
