6J55
 
 | | Crystal structure of an iron superoxide dismutate (FeSOD) from a pathogenic Acanthamoeba castellanii | | Descriptor: | FE (II) ION, Superoxide dismutase | | Authors: | Lee, K.H, Dao, T.O, Asaithambi, K, Na, B.K. | | Deposit date: | 2019-01-10 | | Release date: | 2019-07-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.330005 Å) | | Cite: | Crystal structure of an iron superoxide dismutase from the pathogenic amoeba Acanthamoeba castellanii.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
5FYN
 
 | | Sub-tomogram averaging of Tula virus glycoprotein spike | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, PUUMALA VIRUS GN GLYCOPROTEIN, alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Li, S, Rissanen, I, Zeltina, A, Hepojoki, J, Raghwani, J, Harlos, K, Pybus, O.G, Huiskonen, J.T, Bowden, T.A. | | Deposit date: | 2016-03-08 | | Release date: | 2016-06-08 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (15.6 Å) | | Cite: | A Molecular-Level Account of the Antigenic Hantaviral Surface.
Cell Rep., 15, 2016
|
|
5G13
 
 | |
7BP1
 
 | | Crystal structure of 2, 3-dihydroxybenzoic acid decarboxylase from Fusarium oxysporum in complex with Catechol | | Descriptor: | 2,3-dihydroxybenzoate decarboxylase, CATECHOL, ZINC ION | | Authors: | Song, M.K, Feng, J.H, Liu, W.D, Wu, Q.Q, Zhu, D.M. | | Deposit date: | 2020-03-21 | | Release date: | 2020-07-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | 2,3-Dihydroxybenzoic Acid Decarboxylase from Fusarium oxysporum: Crystal Structures and Substrate Recognition Mechanism.
Chembiochem, 21, 2020
|
|
4QH7
 
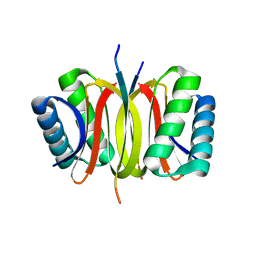 | | LC8 - Ana2 (159-168) Complex | | Descriptor: | Anastral spindle 2, Dynein light chain 1, cytoplasmic | | Authors: | Slevin, L.K, Romes, E.R, Slep, K.C. | | Deposit date: | 2014-05-27 | | Release date: | 2014-06-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.829 Å) | | Cite: | The Mechanism of Dynein Light Chain LC8-mediated Oligomerization of the Ana2 Centriole Duplication Factor.
J.Biol.Chem., 289, 2014
|
|
4QHG
 
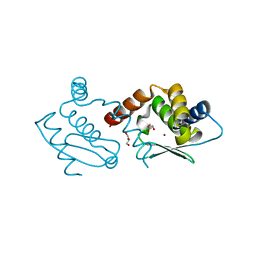 | | Crystal structure of Methanocaldococcus jannaschii dimeric selecase | | Descriptor: | GLYCEROL, Uncharacterized protein MJ1213, ZINC ION | | Authors: | Lopez-pelegrin, M, Cerda-costa, N, Cintas-pedrola, A, Herranz-trillo, F, Bernado, P, Peinado, J.R, Arolas, J.L, Gomis-ruth, F.X. | | Deposit date: | 2014-05-28 | | Release date: | 2014-07-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Multiple stable conformations account for reversible concentration-dependent oligomerization and autoinhibition of a metamorphic metallopeptidase
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
5G0X
 
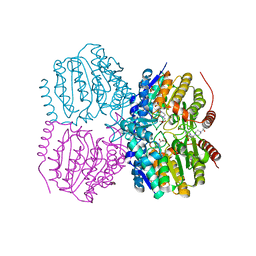 | | Pseudomonas aeruginosa HDAH bound to acetate. | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ACETATE ION, HDAH, ... | | Authors: | Kraemer, A, Meyer-Almes, F.J, Yildiz, O. | | Deposit date: | 2016-03-23 | | Release date: | 2016-11-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of a Histone Deacetylase Homologue from Pseudomonas aeruginosa.
Biochemistry, 55, 2016
|
|
5G10
 
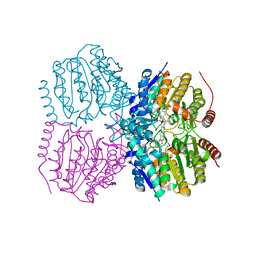 | | Pseudomonas aeruginosa HDAH bound to 9,9,9 trifluoro-8,8-dihydroy-N-phenylnonanamide | | Descriptor: | 9,9,9-tris(fluoranyl)-8,8-bis(oxidanyl)-~{N}-phenyl-nonanamide, HDAH, POTASSIUM ION, ... | | Authors: | Kraemer, A, Meyer-Almes, F.J, Yildiz, O. | | Deposit date: | 2016-03-23 | | Release date: | 2016-11-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Crystal Structure of a Histone Deacetylase Homologue from Pseudomonas aeruginosa.
Biochemistry, 55, 2016
|
|
4QOM
 
 | | Bacillus pumilus catalase with pyrogallol bound | | Descriptor: | BENZENE-1,2,3-TRIOL, CHLORIDE ION, Catalase, ... | | Authors: | Loewen, P.C. | | Deposit date: | 2014-06-20 | | Release date: | 2015-02-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Unprecedented access of phenolic substrates to the heme active site of a catalase: Substrate binding and peroxidase-like reactivity of Bacillus pumilus catalase monitored by X-ray crystallography and EPR spectroscopy.
Proteins, 83, 2015
|
|
1R52
 
 | | Crystal structure of the bifunctional chorismate synthase from Saccharomyces cerevisiae | | Descriptor: | Chorismate synthase | | Authors: | Quevillon-Cheruel, S, Leulliot, N, Meyer, P, Graille, M, Bremang, M, Blondeau, K, Sorel, I, Poupon, A, Janin, J, van Tilbeurgh, H. | | Deposit date: | 2003-10-09 | | Release date: | 2003-12-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Crystal structure of the bifunctional chorismate synthase from Saccharomyces cerevisiae
J.Biol.Chem., 279, 2004
|
|
1FMA
 
 | | MOLYBDOPTERIN SYNTHASE (MOAD/MOAE) | | Descriptor: | CHLORIDE ION, MOLYBDOPTERIN CONVERTING FACTOR, SUBUNIT 1, ... | | Authors: | Rudolph, M.J, Wuebbens, M.M, Rajagolpalan, K.V, Schindelin, H. | | Deposit date: | 2000-08-16 | | Release date: | 2001-01-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystal structure of molybdopterin synthase and its evolutionary relationship to ubiquitin activation.
Nat.Struct.Biol., 8, 2001
|
|
4QOP
 
 | | Structure of Bacillus pumilus catalase with hydroquinone bound. | | Descriptor: | CHLORIDE ION, Catalase, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Loewen, P.C. | | Deposit date: | 2014-06-20 | | Release date: | 2015-02-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Unprecedented access of phenolic substrates to the heme active site of a catalase: Substrate binding and peroxidase-like reactivity of Bacillus pumilus catalase monitored by X-ray crystallography and EPR spectroscopy.
Proteins, 83, 2015
|
|
8Q5O
 
 | |
4QOL
 
 | | Structure of Bacillus pumilus catalase | | Descriptor: | ACETATE ION, CHLORIDE ION, Catalase, ... | | Authors: | Loewen, P.C. | | Deposit date: | 2014-06-20 | | Release date: | 2015-02-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Unprecedented access of phenolic substrates to the heme active site of a catalase: Substrate binding and peroxidase-like reactivity of Bacillus pumilus catalase monitored by X-ray crystallography and EPR spectroscopy.
Proteins, 83, 2015
|
|
2BF7
 
 | | Leishmania major pteridine reductase 1 in complex with NADP and biopterin | | Descriptor: | 1,2-ETHANEDIOL, 7,8-DIHYDROBIOPTERIN, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Schuettelkopf, A.W, Hunter, W.N. | | Deposit date: | 2004-12-06 | | Release date: | 2005-08-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of Leishmania Major Pteridine Reductase Complexes Reveal the Active Site Features Important for Ligand Binding and to Guide Inhibitor Design
J.Mol.Biol., 352, 2005
|
|
4QOQ
 
 | | Structure of Bacillus pumilus catalase with guaiacol bound | | Descriptor: | CHLORIDE ION, Catalase, Guaiacol, ... | | Authors: | Loewen, P.C. | | Deposit date: | 2014-06-20 | | Release date: | 2015-02-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Unprecedented access of phenolic substrates to the heme active site of a catalase: Substrate binding and peroxidase-like reactivity of Bacillus pumilus catalase monitored by X-ray crystallography and EPR spectroscopy.
Proteins, 83, 2015
|
|
4QOO
 
 | | Structure of Bacillus pumilus catalase with resorcinol bound. | | Descriptor: | CHLORIDE ION, Catalase, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Loewen, P.C. | | Deposit date: | 2014-06-20 | | Release date: | 2015-02-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Unprecedented access of phenolic substrates to the heme active site of a catalase: Substrate binding and peroxidase-like reactivity of Bacillus pumilus catalase monitored by X-ray crystallography and EPR spectroscopy.
Proteins, 83, 2015
|
|
4QON
 
 | | Structure of Bacillus pumilus catalase with catechol bound. | | Descriptor: | CATECHOL, CHLORIDE ION, Catalase, ... | | Authors: | Loewen, P.C. | | Deposit date: | 2014-06-20 | | Release date: | 2015-02-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Unprecedented access of phenolic substrates to the heme active site of a catalase: Substrate binding and peroxidase-like reactivity of Bacillus pumilus catalase monitored by X-ray crystallography and EPR spectroscopy.
Proteins, 83, 2015
|
|
2BFM
 
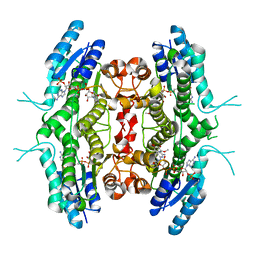 | |
4QOR
 
 | | Structure of Bacillus pumilus catalase with chlorophenol bound. | | Descriptor: | 2-CHLOROPHENOL, CHLORIDE ION, Catalase, ... | | Authors: | Loewen, P.C. | | Deposit date: | 2014-06-20 | | Release date: | 2015-02-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Unprecedented access of phenolic substrates to the heme active site of a catalase: Substrate binding and peroxidase-like reactivity of Bacillus pumilus catalase monitored by X-ray crystallography and EPR spectroscopy.
Proteins, 83, 2015
|
|
2CB1
 
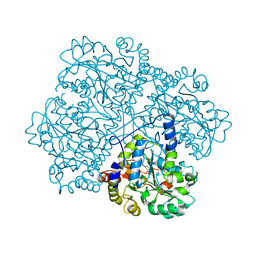 | | Crystal Structure of O-actetyl Homoserine Sulfhydrylase From Thermus Thermophilus HB8,OAH2. | | Descriptor: | O-ACETYL HOMOSERINE SULFHYDRYLASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Imagawa, T, Utsunomiya, H, Tsuge, H, Ebihara, A, Kanagawa, M, Nakagawa, N, Kuroishi, C, Agari, Y, Kuramitsu, S, Yokoyama, S. | | Deposit date: | 2005-12-28 | | Release date: | 2007-01-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Crystal Structure of O-Acetyl Homoserine Sulfhydrylase
To be Published
|
|
6TFJ
 
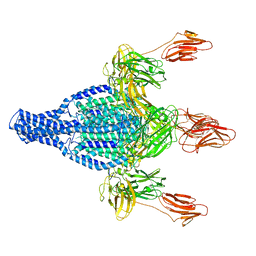 | | Vip3Aa protoxin structure | | Descriptor: | Vegetative insecticidal protein | | Authors: | Nunez-Ramirez, R, Huesa, J, Bel, Y, Ferre, J, Casino, P, Arias-Palomo, E. | | Deposit date: | 2019-11-14 | | Release date: | 2020-08-12 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Molecular architecture and activation of the insecticidal protein Vip3Aa from Bacillus thuringiensis.
Nat Commun, 11, 2020
|
|
5GQQ
 
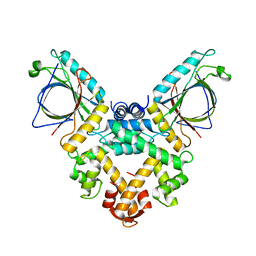 | | Structure of ALG-2/HEBP2 Complex | | Descriptor: | CALCIUM ION, CHLORIDE ION, Heme-binding protein 2, ... | | Authors: | Liu, X, Ma, J, Zhang, H, Feng, Y. | | Deposit date: | 2016-08-08 | | Release date: | 2016-11-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and Functional Study of Apoptosis-linked Gene-2Heme-binding Protein 2 Interactions in HIV-1 Production.
J. Biol. Chem., 291, 2016
|
|
5FXK
 
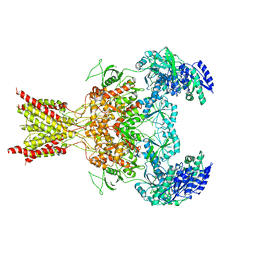 | | GluN1b-GluN2B NMDA receptor structure-Class Y | | Descriptor: | N-METHYL-D-ASPARTATE RECEPTOR GLUN1, N-METHYL-D-ASPARTATE RECEPTOR GLUN2B | | Authors: | Tajima, N, Karakas, E, Grant, T, Simorowski, N, Diaz-Avalos, R, Grigorieff, N, Furukawa, H. | | Deposit date: | 2016-03-02 | | Release date: | 2016-05-11 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (6.4 Å) | | Cite: | Activation of Nmda Receptors and the Mechanism of Inhibition by Ifenprodil.
Nature, 534, 2016
|
|
2BFA
 
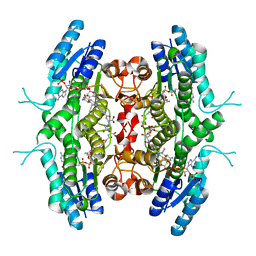 | | Leishmania major pteridine reductase 1 in complex with NADP and CB3717 | | Descriptor: | 1,2-ETHANEDIOL, 10-PROPARGYL-5,8-DIDEAZAFOLIC ACID, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Schuettelkopf, A.W, Hunter, W.N. | | Deposit date: | 2004-12-06 | | Release date: | 2005-08-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structures of Leishmania Major Pteridine Reductase Complexes Reveal the Active Site Features Important for Ligand Binding and to Guide Inhibitor Design
J.Mol.Biol., 352, 2005
|
|
