6YIF
 
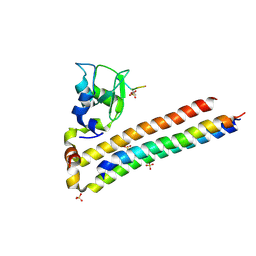 | | Structure of Chromosomal Passenger Complex (CPC) bound to phosphorylated Histone 3 peptide at 1.8 A. | | Descriptor: | Baculoviral IAP repeat-containing protein 5, Borealin, Inner centromere protein, ... | | Authors: | Serena, M, Elliott, P.R, Barr, F.A. | | Deposit date: | 2020-04-01 | | Release date: | 2020-05-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Molecular basis of MKLP2-dependent Aurora B transport from chromatin to the anaphase central spindle.
J.Cell Biol., 219, 2020
|
|
6YIH
 
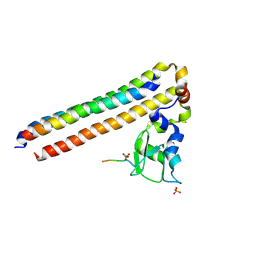 | | Structure of Chromosomal Passenger Complex (CPC) bound to phosphorylated Histone 3 peptide at 2.6 A. | | Descriptor: | Baculoviral IAP repeat-containing protein 5, Borealin, Histone H3.1, ... | | Authors: | Serena, M, Elliott, P.R, Barr, F.A. | | Deposit date: | 2020-04-01 | | Release date: | 2020-05-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Molecular basis of MKLP2-dependent Aurora B transport from chromatin to the anaphase central spindle.
J.Cell Biol., 219, 2020
|
|
3FDT
 
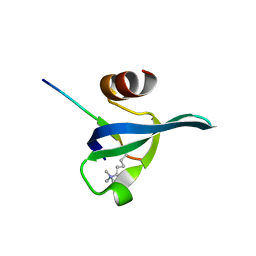 | | Crystal structure of the complex of human chromobox homolog 5 (CBX5) with H3K9(me)3 peptide | | Descriptor: | Chromobox protein homolog 5, H3K9(me)3 peptide | | Authors: | Amaya, M.F, Ravichandran, M, Loppnau, P, Kozieradzki, I, Edwards, A.M, Arrowsmith, C.H, Weigelt, J, Bountra, C, Bochkarev, A, Min, J, Ouyang, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-11-26 | | Release date: | 2009-01-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Recognition and specificity determinants of the human cbx chromodomains.
J.Biol.Chem., 286, 2011
|
|
3GV6
 
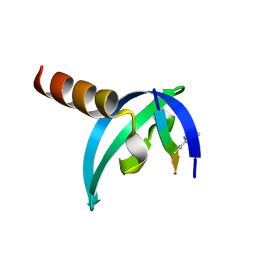 | | Crystal Structure of human chromobox homolog 6 (CBX6) with H3K9 peptide | | Descriptor: | Chromobox protein homolog 6, Histone H3K9me3 peptide | | Authors: | Dong, A, Amaya, M.F, Li, Z, Loppnau, P, Kozieradzki, I, Edwards, A.M, Arrowsmith, C.H, Weigelt, J, Bountra, C, Bochkarev, A, Min, J, Ouyang, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-03-30 | | Release date: | 2009-04-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Recognition and specificity determinants of the human cbx chromodomains.
J.Biol.Chem., 286, 2011
|
|
2VPG
 
 | | Decoding of methylated histone H3 tail by the Pygo-BCL9 Wnt signaling complex | | Descriptor: | B-CELL CLL/LYMPHOMA 9 PROTEIN, GLYCEROL, HISTONE H3 TAIL, ... | | Authors: | Fiedler, M, Sanchez-Barrena, M.J, Nekrasov, M, Mieszczanek, J, Rybin, V, Muller, J, Bienz, M, Evans, P. | | Deposit date: | 2008-02-27 | | Release date: | 2008-06-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Decoding of Methylated Histone H3 Tail by the Pygo- Bcl9 Wnt Signaling Complex.
Mol.Cell, 30, 2008
|
|
2WP2
 
 | | Structure of Brdt bromodomain BD1 bound to a diacetylated histone H4 peptide. | | Descriptor: | BROMODOMAIN TESTIS-SPECIFIC PROTEIN, HISTONE H4 | | Authors: | Moriniere, J, Rousseaux, S, Steuerwald, U, Soler-Lopez, M, Curtet, S, Vitte, A.-L, Govin, J, Gaucher, J, Sadoul, K, Hart, D.J, Krijgsveld, J, Khochbin, S, Mueller, C.W, Petosa, C. | | Deposit date: | 2009-08-02 | | Release date: | 2009-09-22 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Cooperative Binding of Two Acetylation Marks on a Histone Tail by a Single Bromodomain.
Nature, 461, 2009
|
|
2X0L
 
 | | Crystal structure of a neuro-specific splicing variant of human histone lysine demethylase LSD1. | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, HISTONE H3 PEPTIDE, LYSINE-SPECIFIC HISTONE DEMETHYLASE 1, ... | | Authors: | Zibetti, C, Adamo, A, Binda, C, Forneris, F, Verpelli, C, Ginelli, E, Mattevi, A, Sala, C, Battaglioli, E. | | Deposit date: | 2009-12-15 | | Release date: | 2010-03-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Alternative Splicing of the Histone Demethylase Lsd1/Kdm1 Contributes to the Modulation of Neurite Morphogenesis in the Mammalian Nervous System.
J.Neurosci., 30, 2010
|
|
1PEG
 
 | | Structural basis for the product specificity of histone lysine methyltransferases | | Descriptor: | Histone H3, S-ADENOSYL-L-HOMOCYSTEINE, ZINC ION, ... | | Authors: | Zhang, X, Yang, Z, Khan, S.I, Horton, J.R, Tamaru, H, Selker, E.U, Cheng, X. | | Deposit date: | 2003-05-21 | | Release date: | 2003-08-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Structural basis for the product specificity of histone lysine methyltransferases
Mol.Cell, 12, 2003
|
|
2XFM
 
 | | Complex structure of the MIWI Paz domain bound to methylated single stranded RNA | | Descriptor: | 5'-R(*AP*CP*CP*GP*AP*CP*UP*(OMU)P)-3', PIWI-LIKE PROTEIN 1 | | Authors: | Simon, B, Kirkpatrick, J.P, Eckhardt, S, Sehr, P, Andrade-Navarro, M.A, Pillai, R.S, Carlomagno, T. | | Deposit date: | 2010-05-27 | | Release date: | 2011-01-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Recognition of 2'-O-Methylated 3'-End of Pirna by the Paz Domain of a Piwi Protein.
Structure, 19, 2011
|
|
1Q3L
 
 | |
2XL3
 
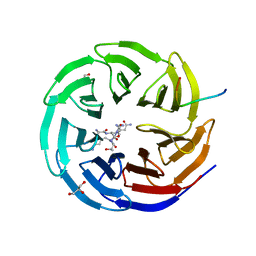 | | WDR5 IN COMPLEX WITH AN RBBP5 PEPTIDE AND HISTONE H3 PEPTIDE | | Descriptor: | GLYCEROL, HISTONE H3.1, RETINOBLASTOMA-BINDING PROTEIN 5, ... | | Authors: | Odho, Z, Southall, S.M, Wilson, J.R. | | Deposit date: | 2010-07-19 | | Release date: | 2010-08-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Characterisation of a Novel Wdr5 Binding Site that Recruits Rbbp5 Through a Conserved Motif and Enhances Methylation of H3K4 by Mll1.
J.Biol.Chem., 285, 2010
|
|
3A1B
 
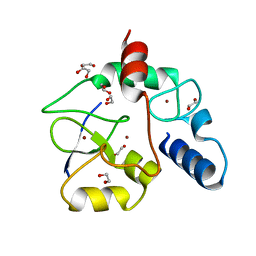 | | Crystal structure of the DNMT3A ADD domain in complex with histone H3 | | Descriptor: | 1,2-ETHANEDIOL, DNA (cytosine-5)-methyltransferase 3A, Histone H3.1, ... | | Authors: | Otani, J, Arita, K, Ariyoshi, M, Shirakawa, M. | | Deposit date: | 2009-03-28 | | Release date: | 2009-11-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.292 Å) | | Cite: | Structural basis for recognition of H3K4 methylation status by the DNA methyltransferase 3A ATRX-DNMT3-DNMT3L domain
Embo Rep., 10, 2009
|
|
3AN2
 
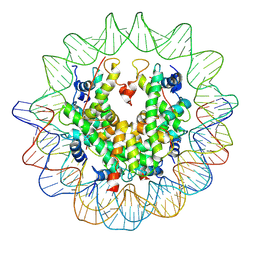 | | The structure of the centromeric nucleosome containing CENP-A | | Descriptor: | 147 mer DNA, Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Tachiwana, H, Kagawa, W, Shiga, T, Saito, K, Osakabe, A, Hayashi-Takanaka, Y, Park, S.-Y, Kimura, H, Kurumizaka, H. | | Deposit date: | 2010-08-27 | | Release date: | 2011-07-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Crystal structure of the human centromeric nucleosome containing CENP-A
Nature, 476, 2011
|
|
3ASK
 
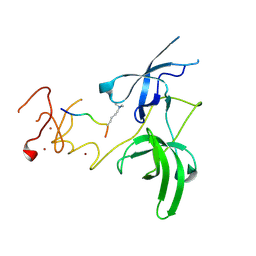 | | Structure of UHRF1 in complex with histone tail | | Descriptor: | E3 ubiquitin-protein ligase UHRF1, Histone H3.3, ZINC ION | | Authors: | Arita, K, Sugita, K, Unoki, M, Hamamoto, R, Sekiyama, N, Tochio, H, Ariyoshi, M, Shirakawa, M. | | Deposit date: | 2010-12-16 | | Release date: | 2012-01-25 | | Last modified: | 2013-06-05 | | Method: | X-RAY DIFFRACTION (2.904 Å) | | Cite: | Recognition of modification status on a histone H3 tail by linked histone reader modules of the epigenetic regulator UHRF1
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3ASL
 
 | | Structure of UHRF1 in complex with histone tail | | Descriptor: | 1,2-ETHANEDIOL, E3 ubiquitin-protein ligase UHRF1, Histone H3.3, ... | | Authors: | Arita, K, Sugita, K, Unoki, M, Hamamoto, R, Sekiyama, N, Tochio, H, Ariyoshi, M, Shirakawa, M. | | Deposit date: | 2010-12-16 | | Release date: | 2012-01-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Recognition of modification status on a histone H3 tail by linked histone reader modules of the epigenetic regulator UHRF1
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3B95
 
 | | EuHMT1 (Glp) Ankyrin Repeat Domain (Structure 2) | | Descriptor: | Euchromatic histone-lysine N-methyltransferase 1, Histone H3 N-terminal Peptide, SULFATE ION | | Authors: | Collins, R.E, Horton, J.R, Cheng, X. | | Deposit date: | 2007-11-02 | | Release date: | 2008-02-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | The ankyrin repeats of G9a and GLP histone methyltransferases are mono- and dimethyllysine binding modules.
Nat.Struct.Mol.Biol., 15, 2008
|
|
7CJ0
 
 | |
7CIZ
 
 | |
7CFB
 
 | | Drosophila melanogaster Krimper eTud1 apo structure | | Descriptor: | FI20010p1, SULFATE ION | | Authors: | Hu, H, Li, S. | | Deposit date: | 2020-06-25 | | Release date: | 2021-06-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Binding of guide piRNA triggers methylation of the unstructured N-terminal region of Aub leading to assembly of the piRNA amplification complex.
Nat Commun, 12, 2021
|
|
7CFC
 
 | | Drosophila melanogaster Krimper eTud1-Ago3 complex | | Descriptor: | FI20010p1, Protein argonaute-3 | | Authors: | Hu, H, Li, S. | | Deposit date: | 2020-06-25 | | Release date: | 2021-06-02 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Binding of guide piRNA triggers methylation of the unstructured N-terminal region of Aub leading to assembly of the piRNA amplification complex.
Nat Commun, 12, 2021
|
|
7CFD
 
 | | Drosophila melanogaster Krimper eTud2-AubR15me2 complex | | Descriptor: | FI20010p1, Protein aubergine | | Authors: | Hu, H, Li, S. | | Deposit date: | 2020-06-25 | | Release date: | 2021-06-02 | | Last modified: | 2021-09-15 | | Method: | X-RAY DIFFRACTION (2.704 Å) | | Cite: | Binding of guide piRNA triggers methylation of the unstructured N-terminal region of Aub leading to assembly of the piRNA amplification complex.
Nat Commun, 12, 2021
|
|
7CFP
 
 | |
7CFQ
 
 | | Crystal structure of WDR5 in complex with H3K4me3Q5ser peptide | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, H3K4me3Q5ser peptide, ... | | Authors: | Zhao, J, Zhang, X, Zang, J. | | Deposit date: | 2020-06-27 | | Release date: | 2021-07-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural insights into the recognition of histone H3Q5 serotonylation by WDR5.
Sci Adv, 7, 2021
|
|
7EBK
 
 | |
7EIC
 
 | |
