3O2K
 
 | | Crystal Structure of Brevianamide F Prenyltransferase Complexed with Brevianamide F and Dimethylallyl S-thiolodiphosphate | | Descriptor: | (3S,8aS)-3-(1H-indol-3-ylmethyl)hexahydropyrrolo[1,2-a]pyrazine-1,4-dione, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Brevianamide F prenyltransferase, ... | | Authors: | Jost, M, Zocher, G.E, Stehle, T. | | Deposit date: | 2010-07-22 | | Release date: | 2010-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-function analysis of an enzymatic prenyl transfer reaction identifies a reaction chamber with modifiable specificity.
J.Am.Chem.Soc., 132, 2010
|
|
3KMZ
 
 | | Crystal structure of RARalpha ligand binding domain in complex with the inverse agonist BMS493 and a corepressor fragment | | Descriptor: | 4-{(E)-2-[5,5-dimethyl-8-(phenylethynyl)-5,6-dihydronaphthalen-2-yl]ethenyl}benzoic acid, GLYCEROL, Nuclear receptor corepressor 1, ... | | Authors: | Bourguet, W, le Maire, A. | | Deposit date: | 2009-11-11 | | Release date: | 2010-06-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A unique secondary-structure switch controls constitutive gene repression by retinoic acid receptor.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3AQT
 
 | | CRYSTAL STRUCTURE OF RolR (NCGL1110) complex WITH ligand RESORCINOL | | Descriptor: | Bacterial regulatory proteins, tetR family, RESORCINOL | | Authors: | Li, D.F, Zhang, N, Hou, Y.J, Liu, S.J, Wang, D.C. | | Deposit date: | 2010-11-18 | | Release date: | 2011-07-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of the transcriptional repressor RolR reveals a novel recognition mechanism between inducer and regulator.
Plos One, 6, 2011
|
|
4E4D
 
 | | Crystal structure of mouse RANKL-OPG complex | | Descriptor: | CHLORIDE ION, Tumor necrosis factor ligand superfamily member 11, soluble form, ... | | Authors: | Nelson, C.A, Fremont, D.H. | | Deposit date: | 2012-03-12 | | Release date: | 2012-10-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | RANKL Employs Distinct Binding Modes to Engage RANK and the Osteoprotegerin Decoy Receptor.
Structure, 20, 2012
|
|
4AWM
 
 | | Influenza strain pH1N1 2009 polymerase subunit PA endonuclease in complex with (-)-epigallocatechin gallate from green tea | | Descriptor: | (2R,3R)-5,7-dihydroxy-2-(3,4,5-trihydroxyphenyl)-3,4-dihydro-2H-chromen-3-yl 3,4,5-trihydroxybenzoate, MANGANESE (II) ION, POLYMERASE PA | | Authors: | Kowalinski, E, Zubieta, C, Wolkerstorfer, A, Szolar, O.H, Ruigrok, R.W, Cusack, S. | | Deposit date: | 2012-06-04 | | Release date: | 2012-08-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Analysis of Specific Metal Chelating Inhibitor Binding to the Endonuclease Domain of Influenza Ph1N1 (2009) Polymerase.
Plos Pathog., 8, 2012
|
|
1EXT
 
 | |
3TZS
 
 | |
2M9P
 
 | |
4K72
 
 | |
4K78
 
 | |
3HFT
 
 | |
4K75
 
 | |
5HZW
 
 | | Crystal structure of the orphan region of human endoglin/CD105 in complex with BMP9 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Growth/differentiation factor 2, Maltose-binding periplasmic protein,Endoglin, ... | | Authors: | Bokhove, M, Saito, T, Jovine, L. | | Deposit date: | 2016-02-03 | | Release date: | 2017-06-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (4.451 Å) | | Cite: | Structural Basis of the Human Endoglin-BMP9 Interaction: Insights into BMP Signaling and HHT1.
Cell Rep, 19, 2017
|
|
7BWO
 
 | | Consensus chitin binding protein | | Descriptor: | Chitin binding beak protein 3 | | Authors: | Mohanram, H, Miserez, A. | | Deposit date: | 2020-04-15 | | Release date: | 2021-04-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of a consensus chitin-binding domain revealed by solution NMR.
J.Struct.Biol., 213, 2021
|
|
7BWE
 
 | | Consensus Chitin binding domain | | Descriptor: | Chitin binding beak protein 3 | | Authors: | Mohanram, H, Miserez, A. | | Deposit date: | 2020-04-14 | | Release date: | 2021-04-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of a consensus chitin-binding domain revealed by solution NMR.
J.Struct.Biol., 213, 2021
|
|
6NM1
 
 | | The crystal structure of the Staphylococcus aureus Fatty acid Kinase (Fak) B1 protein A158L mutant to 2.33 Angstrom resolution exhibits a conformation change compared to the wild type form | | Descriptor: | Fatty acid Kinase (Fak) B1 protein, MYRISTIC ACID | | Authors: | Cuypers, M.G, Gullett, J.M, Subramanian, C, Ericson, M, White, S.W, Rock, C.O. | | Deposit date: | 2019-01-10 | | Release date: | 2020-01-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Identification of structural transitions in bacterial fatty acid binding proteins that permit ligand entry and exit at membranes.
J.Biol.Chem., 298, 2022
|
|
2M9Q
 
 | |
3VRF
 
 | | The crystal structure of hemoglobin from woolly mammoth in the carbonmonoxy forms | | Descriptor: | CARBON MONOXIDE, Hemoglobin subunit alpha, Hemoglobin subunit beta/delta hybrid, ... | | Authors: | Noguchi, H, Campbell, K.L, Ho, C, Park, S.-Y, Tame, J.R.H. | | Deposit date: | 2012-04-09 | | Release date: | 2012-11-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structures of haemoglobin from woolly mammoth in liganded and unliganded states.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
1A15
 
 | | SDF-1ALPHA | | Descriptor: | STROMAL DERIVED FACTOR-1ALPHA, SULFATE ION | | Authors: | Dealwis, C.G, Fernandez, E.J, Lolis, E. | | Deposit date: | 1997-12-22 | | Release date: | 1998-08-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of chemically synthesized [N33A] stromal cell-derived factor 1alpha, a potent ligand for the HIV-1 "fusin" coreceptor.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
4Q6H
 
 | |
5NZ1
 
 | |
4E0U
 
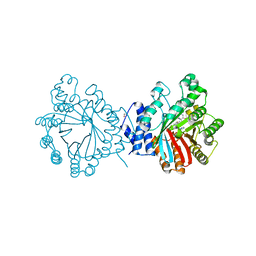 | | Crystal structure of CdpNPT in complex with thiolodiphosphate and (S)-benzodiazependione | | Descriptor: | (3S)-3-(1H-indol-3-ylmethyl)-3,4-dihydro-1H-1,4-benzodiazepine-2,5-dione, 1,2-ETHANEDIOL, Cyclic dipeptide N-prenyltransferase, ... | | Authors: | Schuller, J.M, Zocher, G, Stehle, T. | | Deposit date: | 2012-03-05 | | Release date: | 2012-05-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure and catalytic mechanism of a cyclic dipeptide prenyltransferase with broad substrate promiscuity.
J.Mol.Biol., 422, 2012
|
|
3IEH
 
 | |
4E0T
 
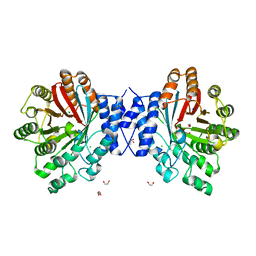 | | Crystal structure of CdpNPT in its unbound state | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Cyclic dipeptide N-prenyltransferase, ... | | Authors: | Schuller, J.M, Zocher, G, Stehle, T. | | Deposit date: | 2012-03-05 | | Release date: | 2012-05-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure and catalytic mechanism of a cyclic dipeptide prenyltransferase with broad substrate promiscuity.
J.Mol.Biol., 422, 2012
|
|
3OJV
 
 | | Crystal Structure of FGF1 complexed with the ectodomain of FGFR1c exhibiting an ordered ligand specificity-determining betaC'-betaE loop | | Descriptor: | 4-deoxy-2-O-sulfo-alpha-L-threo-hex-4-enopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, Basic fibroblast growth factor receptor 1, Heparin-binding growth factor 1 | | Authors: | Beenken, A, Mohammadi, M. | | Deposit date: | 2010-08-23 | | Release date: | 2011-12-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Plasticity in Interactions of Fibroblast Growth Factor 1 (FGF1) N Terminus with FGF Receptors Underlies Promiscuity of FGF1.
J.Biol.Chem., 287, 2012
|
|
