4YNE
 
 | | (R)-2-Phenylpyrrolidine Substitute Imidazopyridazines: a New Class of Potent and Selective Pan-TRK Inhibitors | | Descriptor: | 6-[(2R)-2-(3-fluorophenyl)pyrrolidin-1-yl]-3-(pyridin-2-yl)imidazo[1,2-b]pyridazine, GLYCEROL, High affinity nerve growth factor receptor, ... | | Authors: | Kreusch, A, Rucker, P, Molteni, V, Loren, J. | | Deposit date: | 2015-03-09 | | Release date: | 2015-06-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.0229 Å) | | Cite: | (R)-2-Phenylpyrrolidine Substituted Imidazopyridazines: A New Class of Potent and Selective Pan-TRK Inhibitors.
Acs Med.Chem.Lett., 6, 2015
|
|
7UP2
 
 | | NDM1-inhibitor co-structure | | Descriptor: | (2M)-4'-methyl-2-(2H-tetrazol-5-yl)[1,1'-biphenyl]-3-sulfonamide, Beta-lactamase VIM-1, ZINC ION | | Authors: | Scapin, G, Fischmann, T.O. | | Deposit date: | 2022-04-14 | | Release date: | 2023-03-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Rapid Evolution of a Fragment-like Molecule to Pan-Metallo-Beta-Lactamase Inhibitors: Initial Leads toward Clinical Candidates.
J.Med.Chem., 65, 2022
|
|
7UP1
 
 | | NDM1-inhibitor co-structure | | Descriptor: | (2M)-4'-methyl-2-(2H-tetrazol-5-yl)[1,1'-biphenyl]-3-sulfonamide, CADMIUM ION, Metallo beta-lactamase, ... | | Authors: | Scapin, G, Fischmann, T.O. | | Deposit date: | 2022-04-14 | | Release date: | 2023-03-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.11 Å) | | Cite: | Rapid Evolution of a Fragment-like Molecule to Pan-Metallo-Beta-Lactamase Inhibitors: Initial Leads toward Clinical Candidates.
J.Med.Chem., 65, 2022
|
|
7UP3
 
 | | NDM1-inhibitor co-structure | | Descriptor: | (3P)-4-[4-(hydroxymethyl)phenyl]-3-(2H-tetrazol-5-yl)pyridine-2-sulfonamide, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CADMIUM ION, ... | | Authors: | Scapin, G, Fischmann, T.O. | | Deposit date: | 2022-04-14 | | Release date: | 2023-03-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Rapid Evolution of a Fragment-like Molecule to Pan-Metallo-Beta-Lactamase Inhibitors: Initial Leads toward Clinical Candidates.
J.Med.Chem., 65, 2022
|
|
7UOX
 
 | | NDM1-inhibitor co-structure | | Descriptor: | (2M)-4'-(hydroxymethyl)-2-(1H-tetrazol-5-yl)[1,1'-biphenyl]-3-ol, ACETATE ION, CADMIUM ION, ... | | Authors: | Scapin, G, Fischmann, T.O. | | Deposit date: | 2022-04-14 | | Release date: | 2023-03-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (0.99 Å) | | Cite: | Rapid Evolution of a Fragment-like Molecule to Pan-Metallo-Beta-Lactamase Inhibitors: Initial Leads toward Clinical Candidates.
J.Med.Chem., 65, 2022
|
|
7UOY
 
 | | NDM1-inhibitor co-structure | | Descriptor: | (6P)-4-amino-6-(2H-tetrazol-5-yl)benzene-1,3-disulfonamide, CADMIUM ION, Metallo beta-lactamase, ... | | Authors: | Scapin, G, Fischmann, T.O. | | Deposit date: | 2022-04-14 | | Release date: | 2023-03-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Rapid Evolution of a Fragment-like Molecule to Pan-Metallo-Beta-Lactamase Inhibitors: Initial Leads toward Clinical Candidates.
J.Med.Chem., 65, 2022
|
|
1YAU
 
 | | Structure of Archeabacterial 20S proteasome- PA26 complex | | Descriptor: | GLYCEROL, Proteasome alpha subunit, Proteasome beta subunit, ... | | Authors: | Forster, A, Masters, E.I, Whitby, F.G, Robinson, H, Hill, C.P. | | Deposit date: | 2004-12-17 | | Release date: | 2005-07-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The 1.9 A structure of a proteasome-11S activator complex and implications for proteasome-PAN/PA700 interactions.
Mol.Cell, 18, 2005
|
|
1YAR
 
 | | Structure of Archeabacterial 20S proteasome mutant D9S- PA26 complex | | Descriptor: | GLYCEROL, Proteasome alpha subunit, Proteasome beta subunit, ... | | Authors: | Forster, A, Masters, E.I, Whitby, F.G, Robinson, H, Hill, C.P. | | Deposit date: | 2004-12-17 | | Release date: | 2005-07-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The 1.9 A structure of a proteasome-11S activator complex and implications for proteasome-PAN/PA700 interactions.
Mol.Cell, 18, 2005
|
|
3IAI
 
 | | Crystal structure of the catalytic domain of the tumor-associated human carbonic anhydrase IX | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5-ACETAMIDO-1,3,4-THIADIAZOLE-2-SULFONAMIDE, Carbonic anhydrase 9, ... | | Authors: | Alterio, V, Di Fiore, A, De Simone, G. | | Deposit date: | 2009-07-14 | | Release date: | 2009-09-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the catalytic domain of the tumor-associated human carbonic anhydrase IX.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
6XLO
 
 | | Crystal structure of bRaf in complex with inhibitor | | Descriptor: | 3-(2-cyanopropan-2-yl)-N-[2-fluoro-4-methyl-5-(7-methyl-8-oxo-7,8-dihydropyrido[2,3-d]pyridazin-3-yl)phenyl]benzamide, IODIDE ION, Serine/threonine-protein kinase B-raf | | Authors: | Yin, J, Eigenbrot, C, Wang, W. | | Deposit date: | 2020-06-28 | | Release date: | 2021-05-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.493 Å) | | Cite: | Targeting KRAS Mutant Cancers via Combination Treatment: Discovery of a 5-Fluoro-4-(3 H )-quinazolinone Aryl Urea pan-RAF Kinase Inhibitor.
J.Med.Chem., 64, 2021
|
|
2F9W
 
 | | Structure of the type III CoaA from Pseudomonas aeruginosa | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, PANTOTHENOIC ACID, ... | | Authors: | Leonardi, R, Yun, M.K, Chohnan, S, White, S.W, Rock, C.O, Jackowski, S. | | Deposit date: | 2005-12-06 | | Release date: | 2006-08-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Prokaryotic Type II and Type III Pantothenate Kinases: The Same Monomer Fold Creates Dimers with Distinct Catalytic Properties.
Structure, 14, 2006
|
|
7YJ0
 
 | |
6ADG
 
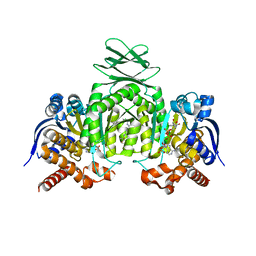 | | Crystal Structures of IDH1 R132H in complex with AG-881 | | Descriptor: | 6-(6-chloropyridin-2-yl)-N2,N4-bis[(2R)-1,1,1-trifluoropropan-2-yl]-1,3,5-triazine-2,4-diamine, Isocitrate dehydrogenase [NADP] cytoplasmic, MAGNESIUM ION, ... | | Authors: | Ma, R, Yun, C.H. | | Deposit date: | 2018-08-01 | | Release date: | 2018-09-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structures of pan-IDH inhibitor AG-881 in complex with mutant human IDH1 and IDH2
Biochem. Biophys. Res. Commun., 503, 2018
|
|
6JBD
 
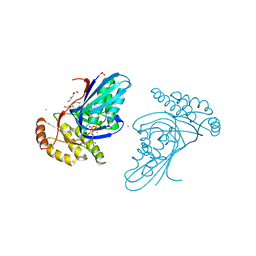 | | Phosphotransferase-ATP complex related to CoA biosynthesis pathway | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, ... | | Authors: | Kita, A, Kishimoto, A, Shimosaka, T, Tomita, H, Yokooji, Y, Imanaka, T, Atomi, H, Miki, K. | | Deposit date: | 2019-01-25 | | Release date: | 2020-01-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of pantoate kinase from Thermococcus kodakarensis.
Proteins, 88, 2020
|
|
7UYA
 
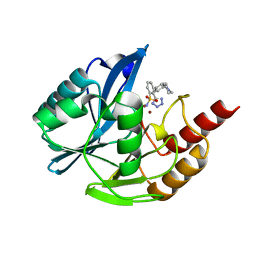 | | Inhibitor bound VIM1 | | Descriptor: | (2M)-4'-(piperidin-4-yl)-2-(1H-tetrazol-5-yl)[1,1'-biphenyl]-3-sulfonamide, Beta-lactamase VIM-1, ZINC ION | | Authors: | Fischmann, T.O, Scapin, G. | | Deposit date: | 2022-05-06 | | Release date: | 2023-05-24 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.01 Å) | | Cite: | Structure Guided Discovery of Novel Pan Metallo-beta-Lactamase Inhibitors with Improved Gram-Negative Bacterial Cell Penetration.
J.Med.Chem., 67, 2024
|
|
7UYD
 
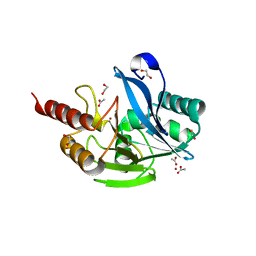 | | Inhibitor bound VIM1 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Metallo-beta-lactamase VIM-2-like protein, ... | | Authors: | Fischmann, T.O, Scapin, G. | | Deposit date: | 2022-05-06 | | Release date: | 2023-05-24 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Structure Guided Discovery of Novel Pan Metallo-beta-Lactamase Inhibitors with Improved Gram-Negative Bacterial Cell Penetration.
J.Med.Chem., 67, 2024
|
|
7UYB
 
 | | Inhibitor bound VIM1 | | Descriptor: | (2M)-4'-(piperidin-4-yl)-2-(1H-tetrazol-5-yl)-4-(trifluoromethyl)[1,1'-biphenyl]-3-sulfonamide, Beta-lactamase VIM-1, ZINC ION | | Authors: | Fischmann, T.O, Scapin, G. | | Deposit date: | 2022-05-06 | | Release date: | 2023-05-24 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.11 Å) | | Cite: | Structure Guided Discovery of Novel Pan Metallo-beta-Lactamase Inhibitors with Improved Gram-Negative Bacterial Cell Penetration.
J.Med.Chem., 67, 2024
|
|
7UYC
 
 | | Inhibitor bound VIM1 | | Descriptor: | (2P)-4'-(piperidin-4-yl)-4-[(piperidin-4-yl)methyl]-2-(1H-tetrazol-5-yl)[1,1'-biphenyl]-3-sulfonamide, Beta-lactamase VIM-1, MAGNESIUM ION, ... | | Authors: | Fischmann, T.O, Scapin, G. | | Deposit date: | 2022-05-06 | | Release date: | 2023-05-24 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | Structure Guided Discovery of Novel Pan Metallo-beta-Lactamase Inhibitors with Improved Gram-Negative Bacterial Cell Penetration.
J.Med.Chem., 67, 2024
|
|
3ET3
 
 | |
5UL1
 
 | | The co-structure of 3-amino-6-(4-((1-(dimethylamino)propan-2-yl)sulfonyl)phenyl)-N-phenylpyrazine-2-carboxamide and a rationally designed PI3K-alpha mutant that mimics ATR | | Descriptor: | 3-amino-6-(4-{[(2S)-1-(dimethylamino)propan-2-yl]sulfonyl}phenyl)-N-phenylpyrazine-2-carboxamide, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Knapp, M.S, Elling, R.A, Mamo, M. | | Deposit date: | 2017-01-23 | | Release date: | 2017-05-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Rationally Designed PI3K alpha Mutants to Mimic ATR and Their Use to Understand Binding Specificity of ATR Inhibitors.
J. Mol. Biol., 429, 2017
|
|
7VR5
 
 | | Crystal structure of CmABCB1 W114Y/W161Y/W363Y/W364Y/M391W (4WY/M391W) mutant | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DECYL-BETA-D-MALTOPYRANOSIDE, Probable ATP-dependent transporter ycf16 | | Authors: | Inoue, Y, Ogawa, H, Kato, H. | | Deposit date: | 2021-10-21 | | Release date: | 2022-09-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure-based alteration of tryptophan residues of the multidrug transporter CmABCB1 to assess substrate binding using fluorescence spectroscopy.
Protein Sci., 31, 2022
|
|
6X5N
 
 | | Crystal structure of a stabilized PAN ENE bimolecular triplex with a GC-clamped polyA tail, in complex with Fab-BL-3,6 | | Descriptor: | Heavy chain Fab Bl-3 6, Light chain Fab BL-3 6, SULFATE ION, ... | | Authors: | Swain, M, Li, M, Woldawer, A, LeGrice, S.F.J. | | Deposit date: | 2020-05-26 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Dynamic bulge nucleotides in the KSHV PAN ENE triple helix provide a unique binding platform for small molecule ligands.
Nucleic Acids Res., 49, 2021
|
|
6X5M
 
 | | Crystal structure of a stabilized PAN ENE bimolecular triplex with a GC-clamped polyA tail, in complex with Fab-BL-3,6. | | Descriptor: | Heavy chain Fab Bl-3 6, Light chain Fab BL-3 6, SULFATE ION, ... | | Authors: | Swain, M, Li, M, Wlodawer, A, Le Grice, S.F.J. | | Deposit date: | 2020-05-26 | | Release date: | 2020-06-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Dynamic bulge nucleotides in the KSHV PAN ENE triple helix provide a unique binding platform for small molecule ligands.
Nucleic Acids Res., 49, 2021
|
|
7C53
 
 | |
6ADI
 
 | | Crystal Structures of IDH2 R140Q in complex with AG-881 | | Descriptor: | 6-(6-chloropyridin-2-yl)-N2,N4-bis[(2R)-1,1,1-trifluoropropan-2-yl]-1,3,5-triazine-2,4-diamine, Isocitrate dehydrogenase [NADP], mitochondrial, ... | | Authors: | Ma, R, Yun, C.H. | | Deposit date: | 2018-08-01 | | Release date: | 2018-09-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.969 Å) | | Cite: | Crystal structures of pan-IDH inhibitor AG-881 in complex with mutant human IDH1 and IDH2
Biochem. Biophys. Res. Commun., 503, 2018
|
|
