4GWF
 
 | | Crystal structure of the tyrosine phosphatase SHP-2 with Y279C mutation | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Qiu, W, Lin, A, Hutchinson, A, Romanov, V, Ruzanov, M, Thompson, C, Lam, K, Kisselman, G, Battaile, K, Chirgadze, N.Y. | | Deposit date: | 2012-09-03 | | Release date: | 2013-09-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the tyrosine phosphatase SHP-2 with Y279C mutation
TO BE PUBLISHED
|
|
5H93
 
 | |
5H98
 
 | |
2VLO
 
 | | K97A mutant of E9 DNase domain in complex with Im9 | | Descriptor: | COLICIN E9, COLICIN-E9 IMMUNITY PROTEIN, SULFATE ION | | Authors: | Keeble, A.H, Joachimiak, L.A, Mate, M.J, Meenan, N, Kirkpatrick, N, Baker, D, Kleanthous, C. | | Deposit date: | 2008-01-15 | | Release date: | 2008-05-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Experimental and Computational Analyses of the Energetic Basis for Dual Recognition of Immunity Proteins by Colicin Endonucleases.
J.Mol.Biol., 379, 2008
|
|
5X9K
 
 | |
7YCF
 
 | | HYDROXYNITRILE LYASE FROM THE MILLIPEDE, Oxidus gracilis IN ACETONITRILE | | Descriptor: | 2-HYDROXY-2-METHYLPROPANENITRILE, CHLORIDE ION, Hydroxynitrile lyase, ... | | Authors: | Chaikaew, S, Watanabe, Y, Zheng, D, Motojima, F, Asano, Y. | | Deposit date: | 2022-07-01 | | Release date: | 2024-01-24 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structure-Based Site-Directed Mutagenesis of Hydroxynitrile Lyase from Cyanogenic Millipede, Oxidus gracilis for Hydrocyanation and Henry Reactions.
Chembiochem, 25, 2024
|
|
7YCB
 
 | | HYDROXYNITRILE LYASE FROM THE MILLIPEDE | | Descriptor: | CHLORIDE ION, GLYCEROL, Hydroxynitrile lyase, ... | | Authors: | Chaikaew, S, Watanabe, Y, Zheng, D, Motojima, F, Asano, Y. | | Deposit date: | 2022-07-01 | | Release date: | 2024-01-24 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structure-Based Site-Directed Mutagenesis of Hydroxynitrile Lyase from Cyanogenic Millipede, Oxidus gracilis for Hydrocyanation and Henry Reactions.
Chembiochem, 25, 2024
|
|
7YCT
 
 | | HYDROXYNITRILE LYASE FROM THE MILLIPEDE, Oxidus gracilis complexed with (R)-2-Chloromandelonitrile | | Descriptor: | (2~{R})-2-(2-chlorophenyl)-2-oxidanyl-ethanenitrile, GLYCEROL, Hydroxynitrile lyase, ... | | Authors: | Chaikaew, S, Watanabe, Y, Zheng, D, Motojima, F, Asano, Y. | | Deposit date: | 2022-07-01 | | Release date: | 2024-01-24 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structure-Based Site-Directed Mutagenesis of Hydroxynitrile Lyase from Cyanogenic Millipede, Oxidus gracilis for Hydrocyanation and Henry Reactions.
Chembiochem, 25, 2024
|
|
7YAX
 
 | | HYDROXYNITRILE LYASE FROM THE MILLIPEDE, | | Descriptor: | CHLORIDE ION, Hydroxynitrile lyase, SULFATE ION | | Authors: | Chaikaew, S, Watanabe, Y, Zheng, D, Motojima, F, Asano, Y. | | Deposit date: | 2022-06-28 | | Release date: | 2024-01-17 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structure-Based Site-Directed Mutagenesis of Hydroxynitrile Lyase from Cyanogenic Millipede, Oxidus gracilis for Hydrocyanation and Henry Reactions.
Chembiochem, 25, 2024
|
|
7YCD
 
 | | HYDROXYNITRILE LYASE FROM THE MILLIPEDE, Oxidus gracilis bound with (R)-(+)-ALPHA-HYDROXYBENZENE-ACETONITRILE | | Descriptor: | (2R)-hydroxy(phenyl)ethanenitrile, Hydroxynitrile lyase, SULFATE ION | | Authors: | Chaikaew, S, Watanabe, Y, Zheng, D, Motojima, F, Asano, Y. | | Deposit date: | 2022-07-01 | | Release date: | 2024-01-24 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structure-Based Site-Directed Mutagenesis of Hydroxynitrile Lyase from Cyanogenic Millipede, Oxidus gracilis for Hydrocyanation and Henry Reactions.
Chembiochem, 25, 2024
|
|
2XXW
 
 | | Crystal structure of the GluK2 (GluR6) D776K LBD dimer in complex with glutamate | | Descriptor: | CHLORIDE ION, GLUTAMATE RECEPTOR, IONOTROPIC KAINATE 2, ... | | Authors: | Nayeem, N, Mayans, O, Green, T. | | Deposit date: | 2010-11-12 | | Release date: | 2011-02-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Conformational Flexibility of the Ligand-Binding Domain Dimer in Kainate Receptor Gating and Desensitization
J.Neurosci., 31, 2011
|
|
3F92
 
 | | Crystal structure of ubiquitin-conjugating enzyme E2-25kDa (Huntington Interacting Protein 2) M172A mutant crystallized at pH 8.5 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BETA-MERCAPTOETHANOL, CALCIUM ION, ... | | Authors: | Wilson, R.C, Hughes, R.C, Flatt, J.W, Meehan, E.J, Ng, J.D, Twigg, P.D. | | Deposit date: | 2008-11-13 | | Release date: | 2008-11-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Structure of full-length ubiquitin-conjugating enzyme E2-25K (huntingtin-interacting protein 2).
Acta Crystallogr.,Sect.F, 65, 2009
|
|
7ZBP
 
 | | Unspecific peroxygenase from Marasmius rotula | | Descriptor: | ACETATE ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Santillana, E, Romero, A. | | Deposit date: | 2022-03-24 | | Release date: | 2022-05-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural Characterization of Two Short Unspecific Peroxygenases: Two Different Dimeric Arrangements.
Antioxidants, 11, 2022
|
|
7ZCL
 
 | | Unspecific peroxygenase from Collariella virescens | | Descriptor: | Collariella virescens UPO, HEME C, MAGNESIUM ION | | Authors: | Santillana, E, Romero, A. | | Deposit date: | 2022-03-28 | | Release date: | 2022-05-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Characterization of Two Short Unspecific Peroxygenases: Two Different Dimeric Arrangements.
Antioxidants, 11, 2022
|
|
3U0N
 
 | |
3O9T
 
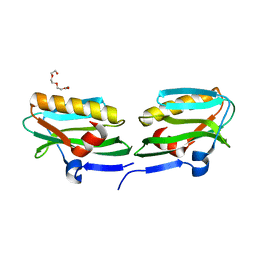 | | Effector domain from influenza A/PR/8/34 NS1 | | Descriptor: | HEXAETHYLENE GLYCOL, Nonstructural protein 1 | | Authors: | Kerry, P.S, Lewis, A, Hale, B.G, Hass, C, Taylor, M.A, Randall, R.E, Russell, R.J.M. | | Deposit date: | 2010-08-04 | | Release date: | 2011-05-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A Transient Homotypic Interaction Model for the Influenza A Virus NS1 Protein Effector Domain.
Plos One, 6, 2011
|
|
3O9U
 
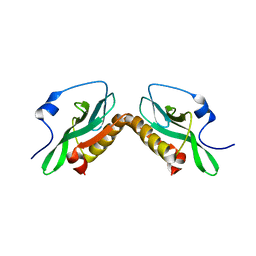 | | Effector domain of influenza A/PR/8/34 NS1 | | Descriptor: | Nonstructural protein 1 | | Authors: | Kerry, P.S, Lewis, A, Hale, B.G, Hass, C, Taylor, M.A, Randall, R.E, Russell, R.J.M. | | Deposit date: | 2010-08-04 | | Release date: | 2011-05-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | A Transient Homotypic Interaction Model for the Influenza A Virus NS1 Protein Effector Domain.
Plos One, 6, 2011
|
|
7Z2P
 
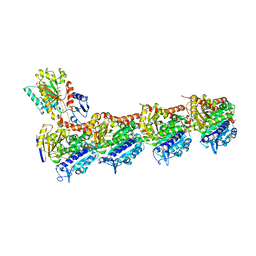 | | Tubulin-nocodazole complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Prota, A.E, Muehlethaler, T, Cavalli, A, Steinmetz, M.O. | | Deposit date: | 2022-02-28 | | Release date: | 2022-08-31 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Novel fragment-derived colchicine-site binders as microtubule-destabilizing agents.
Eur.J.Med.Chem., 241, 2022
|
|
2Y8R
 
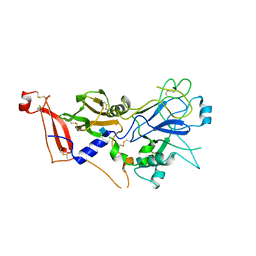 | | Crystal structure of apo AMA1 mutant (Tyr230Ala) from Toxoplasma gondii | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, APICAL MEMBRANE ANTIGEN, PUTATIVE | | Authors: | Tonkin, M.L, Roques, M, Lamarque, M.H, Pugniere, M, Douguet, D, Crawford, J, Lebrun, M, Boulanger, M.J. | | Deposit date: | 2011-02-10 | | Release date: | 2011-08-03 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Host Cell Invasion by Apicomplexan Parasites: Insights from the Co-Structure of Ama1 with a Ron2 Peptide
Science, 333, 2011
|
|
7Z2N
 
 | | Tubulin-18-complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Muehlethaler, T, Prota, A.E, Cavalli, A, Steinmetz, M.O. | | Deposit date: | 2022-02-28 | | Release date: | 2022-08-31 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Novel fragment-derived colchicine-site binders as microtubule-destabilizing agents.
Eur.J.Med.Chem., 241, 2022
|
|
7ZC2
 
 | | Dipeptide and tripeptide Permease C (DtpC) | | Descriptor: | Amino acid/peptide transporter | | Authors: | Killer, M, Finocchio, G, Pardon, E, Steyaert, J, Loew, C. | | Deposit date: | 2022-03-25 | | Release date: | 2022-07-06 | | Last modified: | 2022-08-10 | | Method: | ELECTRON MICROSCOPY (2.72 Å) | | Cite: | Cryo-EM Structure of an Atypical Proton-Coupled Peptide Transporter: Di- and Tripeptide Permease C.
Front Mol Biosci, 9, 2022
|
|
3QDP
 
 | |
3QFQ
 
 | |
2Y7M
 
 | |
7ZES
 
 | | Human SLFN11 dimer bound to ssDNA | | Descriptor: | DNA (5'-D(P*CP*GP*CP*GP*T)-3'), MAGNESIUM ION, Schlafen family member 11, ... | | Authors: | Metzner, F.J, Kugler, M, Wenzl, S.J, Lammens, K. | | Deposit date: | 2022-03-31 | | Release date: | 2022-09-28 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Mechanistic understanding of human SLFN11.
Nat Commun, 13, 2022
|
|
