5POW
 
 | | PanDDA analysis group deposition -- Crystal Structure of BRD1 in complex with N10894b | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 1, N-[(pyridin-2-yl)methyl]acetamide, ... | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-07 | | Release date: | 2017-03-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.768 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
4GWQ
 
 | | Structure of the Mediator Head Module from S. cerevisiae in complex with the carboxy-terminal domain (CTD) of RNA Polymerase II Rpb1 subunit | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, Mediator of RNA polymerase II transcription subunit 11, Mediator of RNA polymerase II transcription subunit 17, ... | | Authors: | Robinson, P.J.J, Bushnell, D.A, Trnka, M.J, Burlingame, A.L, Kornberg, R.D. | | Deposit date: | 2012-09-03 | | Release date: | 2012-10-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (4.5 Å) | | Cite: | Structure of the Mediator Head module bound to the carboxy-terminal domain of RNA polymerase II.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
1YHZ
 
 | | Crystal structure of Arabidopsis thaliana Acetohydroxyacid synthase In Complex With A Sulfonylurea Herbicide, Chlorsulfuron | | Descriptor: | 1-(2-CHLOROPHENYLSULFONYL)-3-(4-METHOXY-6-METHYL-L,3,5-TRIAZIN-2-YL)UREA, 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Acetolactate synthase, ... | | Authors: | McCourt, J.A, Pang, S.S, King-Scott, J, Guddat, L.W, Duggleby, R.G. | | Deposit date: | 2005-01-10 | | Release date: | 2006-01-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Herbicide-binding sites revealed in the structure of plant acetohydroxyacid synthase
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
3NBY
 
 | | Crystal structure of the PKI NES-CRM1-RanGTP nuclear export complex | | Descriptor: | Exportin-1, GTP-binding nuclear protein Ran, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Guttler, T, Madl, T, Neumann, P, Deichsel, D, Corsini, L, Monecke, T, Ficner, R, Sattler, M, Gorlich, D. | | Deposit date: | 2010-06-04 | | Release date: | 2010-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.42 Å) | | Cite: | NES consensus redefined by structures of PKI-type and Rev-type nuclear export signals bound to CRM1.
Nat.Struct.Mol.Biol., 17, 2010
|
|
7BBG
 
 | | CRYSTAL STRUCTURE OF HLA-A2-WT1-RMF AND FAB 11D06 | | Descriptor: | Beta-2-microglobulin, Heavy chain of Fab fragment 11D06, Light chain of Fab fragment 11D06, ... | | Authors: | Bujotzek, A, Georges, G, Hanisch, L.J, Klein, C, Benz, J. | | Deposit date: | 2020-12-17 | | Release date: | 2021-10-27 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Targeting intracellular WT1 in AML with a novel RMF-peptide-MHC-specific T-cell bispecific antibody.
Blood, 138, 2021
|
|
3I5V
 
 | |
7BED
 
 | |
5OCV
 
 | | A Rare Lysozyme Crystal Form Solved Using High-Redundancy 3D Electron Diffraction Data from Micron-Sized Needle Shaped Crystals | | Descriptor: | Lysozyme C, SODIUM ION | | Authors: | Xu, H, Lebrette, H, Yang, T, Srinivas, V, Hovmoller, S, Hogbom, M, Zou, X. | | Deposit date: | 2017-07-03 | | Release date: | 2018-03-28 | | Last modified: | 2024-01-17 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.2 Å) | | Cite: | A Rare Lysozyme Crystal Form Solved Using Highly Redundant Multiple Electron Diffraction Datasets from Micron-Sized Crystals.
Structure, 26, 2018
|
|
3HTQ
 
 | | the hemagglutinin structure of an avian H1N1 influenza A virus in complex with LSTc | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, ... | | Authors: | Wang, G, Li, A, Zhang, Q, Wu, C, Zhang, R, Cai, Q, Song, W, Yuen, K.-Y. | | Deposit date: | 2009-06-12 | | Release date: | 2009-08-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.955 Å) | | Cite: | The hemagglutinin structure of an avian H1N1 influenza A virus
Virology, 392, 2009
|
|
3HUJ
 
 | |
7BGU
 
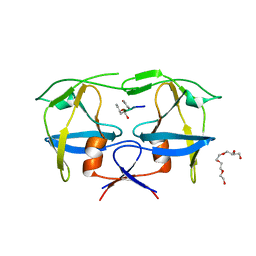 | | Mason-Pfizer Monkey Virus Protease mutant C7A/D26N/C106A in complex with peptidomimetic inhibitor | | Descriptor: | Gag-Pro-Pol polyprotein, PENTAETHYLENE GLYCOL, peptidomimetic inhibitor | | Authors: | Wosicki, S, Gilski, M, Kazmierczyk, M, Jaskolski, M, Zabranska, H, Pichova, I. | | Deposit date: | 2021-01-08 | | Release date: | 2021-12-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.433 Å) | | Cite: | Crystal structures of inhibitor complexes of M-PMV protease with visible flap loops.
Protein Sci., 30, 2021
|
|
3HZL
 
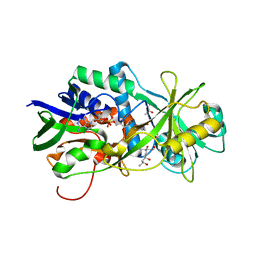 | | Tyr258Phe mutant of NikD, an unusual amino acid oxidase essential for nikkomycin biosynthesis: open form at 1.55A resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, FLAVIN-ADENINE DINUCLEOTIDE, NikD protein, ... | | Authors: | Mathews, F.S, Jorns, M.S, Carrell, C.J. | | Deposit date: | 2009-06-23 | | Release date: | 2009-10-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Factors that affect oxygen activation and coupling of the two redox cycles in the aromatization reaction catalyzed by NikD, an unusual amino acid oxidase.
Biochemistry, 48, 2009
|
|
1XOE
 
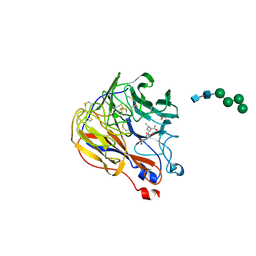 | | N9 Tern influenza neuraminidase complexed with (2R,4R,5R)-5-(1-Acetylamino-3-methyl-butyl-pyrrolidine-2, 4-dicarobyxylic acid 4-methyl esterdase complexed with | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-[1-(ACETYLAMINO)-3-METHYLBUTYL]-4-(METHOXYCARBONYL)PROLINE, Neuraminidase, ... | | Authors: | Wang, G.T, Wang, S, Gentles, R, Sowin, T, Maring, C.J, Kempf, D.J, Kati, W.M, Stoll, V, Stewart, K.D, Laver, G. | | Deposit date: | 2004-10-06 | | Release date: | 2005-01-11 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Design, synthesis, and structural analysis of inhibitors of influenza neuraminidase
containing a 2,3-disubstituted tetrahydrofuran-5-carboxylic acid core.
Bioorg.Med.Chem.Lett., 15, 2005
|
|
1XQ6
 
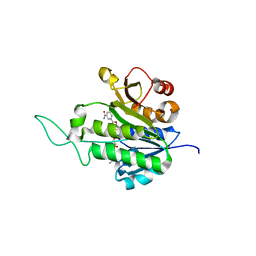 | | X-ray Structure of Gene Product from Arabidopsis Thaliana At5g02240 | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, unknown protein | | Authors: | Wesenberg, G.E, Smith, D.W, Phillips Jr, G.N, Bitto, E, Bingman, C.A, Allard, S.T.M, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2004-10-11 | | Release date: | 2004-10-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray Structure of Gene Product from Arabidopsis Thaliana At5g02240
To be published
|
|
5O05
 
 | |
7B8V
 
 | |
5O02
 
 | |
5NUR
 
 | | Structural basis for maintenance of bacterial outer membrane lipid asymmetry | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, 3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid-(2-4)-3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid-(2-6)-2-amino-2-deoxy-4-O-phosphono-beta-D-glucopyranose-(1-6)-2-amino-2-deoxy-1-O-phosphono-alpha-D-glucopyranose, 3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid-(3-4)-3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid-(2-6)-2-amino-2-deoxy-4-O-phosphono-beta-D-glucopyranose-(1-6)-2-amino-2-deoxy-1-O-phosphono-alpha-D-glucopyranose, ... | | Authors: | Abellon-Ruiz, J, Kaptan, S.S, Basle, A, Claudi, B, Bumann, D, Kleinekathofer, U, van den Berg, B. | | Deposit date: | 2017-05-01 | | Release date: | 2017-10-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.29 Å) | | Cite: | Structural basis for maintenance of bacterial outer membrane lipid asymmetry.
Nat Microbiol, 2, 2017
|
|
3IY9
 
 | | Leishmania Tarentolae Mitochondrial Large Ribosomal Subunit Model | | Descriptor: | 39S ribosomal protein L11, mitochondrial, 39S ribosomal protein L16, ... | | Authors: | Sharma, M.R, Booth, T.M, Simpson, L, Maslov, D.A, Agrawal, R.K. | | Deposit date: | 2009-04-20 | | Release date: | 2009-07-07 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (14.1 Å) | | Cite: | Structure of a mitochondrial ribosome with minimal RNA
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3J16
 
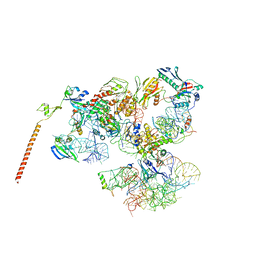 | | Models of ribosome-bound Dom34p and Rli1p and their ribosomal binding partners | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S24-A, ... | | Authors: | Becker, T, Franckenberg, S, Wickles, S, Shoemaker, C.J, Anger, A.M, Armache, J.-P, Sieber, H, Ungewickell, C, Berninghausen, O, Daberkow, I, Karcher, A, Thomm, M, Hopfner, K.-P, Green, R, Beckmann, R. | | Deposit date: | 2011-12-12 | | Release date: | 2012-02-22 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (7.2 Å) | | Cite: | Structural basis of highly conserved ribosome recycling in eukaryotes and archaea.
Nature, 482, 2012
|
|
5NYV
 
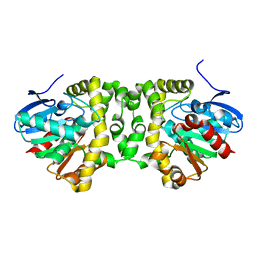 | | Crystal structure determination from picosecond infrared laser ablated protein crystals by serial synchrotron crystallography | | Descriptor: | Fluoroacetate dehalogenase | | Authors: | Schulz, E.C, Kaub, J, Busse, F, Mehrabi, P, Mueller-Werkmeiser, H, Pai, E.F, Robertson, W.D, Miller, R.J.D. | | Deposit date: | 2017-05-11 | | Release date: | 2018-03-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Protein crystals IR laser ablated from aqueous solution at high speed retain their diffractive properties: applications in high-speed serial crystallography.
J.Appl.Crystallogr., 50, 2017
|
|
5O04
 
 | |
1XZW
 
 | | Sweet potato purple acid phosphatase/phosphate complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, FE (III) ION, ... | | Authors: | Schenk, G, Carrington, L.E, Gahan, L.R, Hamilton, S.E, de Jersey, J, Guddat, L.W. | | Deposit date: | 2004-11-12 | | Release date: | 2004-12-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Phosphate forms an unusual tripodal complex with the Fe-Mn center of sweet potato purple acid phosphatase
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
4I0C
 
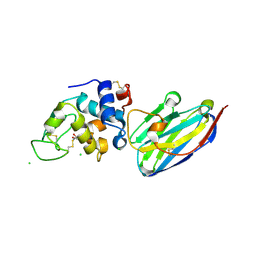 | | The structure of the camelid antibody cAbHuL5 in complex with human lysozyme | | Descriptor: | CHLORIDE ION, GLYCEROL, Lysozyme C, ... | | Authors: | De Genst, E, Chan, P.H, Pardon, E, Kumita, J.R, Christodoulou, J, Menzer, L, Chirgadze, D.Y, Robinson, C.V, Muyldermans, S, Matagne, A, Wyns, L, Dobson, C.M, Dumoulin, M. | | Deposit date: | 2012-11-16 | | Release date: | 2013-10-09 | | Last modified: | 2013-11-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A nanobody binding to non-amyloidogenic regions of the protein human lysozyme enhances partial unfolding but inhibits amyloid fibril formation.
J.Phys.Chem.B, 117, 2013
|
|
4PSO
 
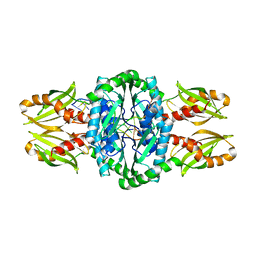 | | Crystal structure of apeThermo-DBP-RP2 bound to ssDNA dT10 | | Descriptor: | PHOSPHATE ION, polydeoxyribonucleotide, ssDNA binding protein | | Authors: | Gahlei, H, von Moeller, H, Eppers, D, Loll, B, Wahl, M.C. | | Deposit date: | 2014-03-07 | | Release date: | 2014-04-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Entrapment of DNA in an intersubunit tunnel system of a single-stranded DNA-binding protein.
Nucleic Acids Res., 42, 2014
|
|
