8EVO
 
 | |
8ET0
 
 | | Crystal Complex of murine Cyclooxygenase-2 with alpaca nanobody F9 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, IBUPROFEN, ... | | Authors: | Xu, S, Banerjee, S, Uddin, M.J, Goodman, M.C, Marnett, L.J. | | Deposit date: | 2022-10-15 | | Release date: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal complex of murine cycloxygenase-2 with alpaca nanobody F9
To Be Published
|
|
8EO6
 
 | | Crystal structure of metagenomic class A beta-lactamase precursor LRA-5 in complex with ceftazidime at 2.35 Angstrom resolution | | Descriptor: | ACYLATED CEFTAZIDIME, LRA-5 | | Authors: | Power, P, D'Amico Gonzalez, G, Centron, D, Gutkind, G, Handelsman, J, Klinke, S. | | Deposit date: | 2022-10-02 | | Release date: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Playing beta-Lactamase Evolution: Metagenomic Class A beta-Lactamase LRA-5 is an Inactive Enzyme Capable of Rendering an Active beta-Lactamase by Introduction of Y69Q and V166E Substitutions
to be published
|
|
8EI7
 
 | | Crystal structure of the WWP2 HECT domain in complex with H304, a Helicon Polypeptide | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Li, K, Tokareva, O.S, Thomson, T.M, Verdine, G.L, McGee, J.H. | | Deposit date: | 2022-09-14 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Recognition and reprogramming of E3 ubiquitin ligase surfaces by alpha-helical peptides.
Nat Commun, 14, 2023
|
|
8EI2
 
 | | Crystal structure of the N-terminal domain of CUL5 in complex with H314, a Helicon Polypeptide | | Descriptor: | Cullin-5, H314, N,N'-(1,4-phenylene)diacetamide | | Authors: | Li, K, Tokareva, O.S, Thomson, T.M, Verdine, G.L, McGee, J.H. | | Deposit date: | 2022-09-14 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Recognition and reprogramming of E3 ubiquitin ligase surfaces by alpha-helical peptides.
Nat Commun, 14, 2023
|
|
8OKJ
 
 | | Carbonic Anhydrase IX like mutant in Complex with Steriod_Sulphamoyl inhibitor AKI_12 | | Descriptor: | Carbonic anhydrase 2, ZINC ION, [(3~{R},5~{S},8~{R},9~{S},10~{S},13~{S},14~{S},17~{S})-17-ethanoyl-10,13-dimethyl-2,3,4,5,6,7,8,9,11,12,14,15,16,17-tetradecahydro-1~{H}-cyclopenta[a]phenanthren-3-yl] sulfamate | | Authors: | Brynda, J, Rezacova, P.M, Kudova, E. | | Deposit date: | 2023-03-28 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Carbonic Anhydrase II in Complex with Steriod_Sulphamoyl
To Be Published
|
|
5LFX
 
 | |
8EIC
 
 | | Crystal structure of beta-catenin and the MDM2 p53-binding domain in complex with H330, a Helicon Polypeptide | | Descriptor: | Catenin beta-1, E3 ubiquitin-protein ligase Mdm2, H330, ... | | Authors: | Li, K, Travaline, T.L, Swiecicki, J.-M, Tokareva, O.S, Thomson, T.M, Verdine, G.L, McGee, J.H. | | Deposit date: | 2022-09-14 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Recognition and reprogramming of E3 ubiquitin ligase surfaces by alpha-helical peptides.
Nat Commun, 14, 2023
|
|
8OLF
 
 | | Carbonic Anhydrase IX like mutant in Complex with Steriod_Sulphamoyl inhibitor BS1982 | | Descriptor: | Carbonic anhydrase 2, ZINC ION, [(8~{S},9~{S},13~{S},14~{S})-13-methyl-6,7,8,9,11,12,14,15,16,17-decahydrocyclopenta[a]phenanthren-3-yl] sulfamate | | Authors: | Brynda, J, Rezacova, P.M, Kudova, E. | | Deposit date: | 2023-03-30 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Carbonic Anhydrase II in Complex with Steriod_Sulphamoyl
To Be Published
|
|
6XKR
 
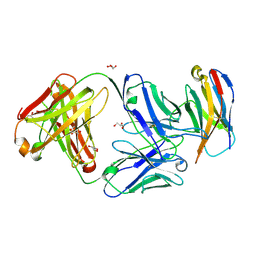 | | Structure of Sasanlimab Fab in complex with PD-1 | | Descriptor: | GLYCEROL, Programmed cell death protein 1, Sasanlimab Fab Heavy chain, ... | | Authors: | Kimberlin, C.R, Chin, S.M. | | Deposit date: | 2020-06-27 | | Release date: | 2020-09-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Pharmacologic Properties and Preclinical Activity of Sasanlimab, A High-affinity Engineered Anti-Human PD-1 Antibody.
Mol.Cancer Ther., 19, 2020
|
|
8EIB
 
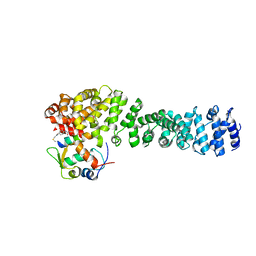 | | Crystal structure of beta-catenin and the MDM2 p53-binding domain in complex with H329, a Helicon Polypeptide | | Descriptor: | Catenin beta-1, E3 ubiquitin-protein ligase Mdm2, H329, ... | | Authors: | Li, K, Travaline, T.L, Swiecicki, J.-M, Tokareva, O.S, Thomson, T.M, Verdine, G.L, McGee, J.H. | | Deposit date: | 2022-09-14 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.76 Å) | | Cite: | Recognition and reprogramming of E3 ubiquitin ligase surfaces by alpha-helical peptides.
Nat Commun, 14, 2023
|
|
8OLP
 
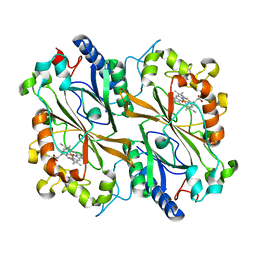 | |
5LHA
 
 | |
8EL5
 
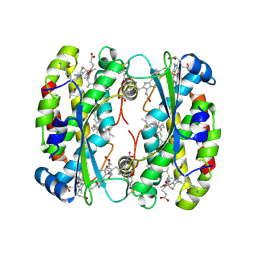 | | Light harvesting phycobiliprotein HaPE555 from the cryptophyte Hemiselmis andersenii CCMP644 in an alternating tight to loose interface filament | | Descriptor: | DiCys-(15,16)-Dihydrobiliverdin, GLYCEROL, PHYCOERYTHROBILIN, ... | | Authors: | Rathbone, H.W, Michie, K.A, Laos, A.L, Curmi, P.M.G. | | Deposit date: | 2022-09-23 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Molecular dissection of the soluble photosynthetic antenna from the cryptophyte alga Hemiselmis andersenii.
Commun Biol, 6, 2023
|
|
8EL4
 
 | | Light harvesting phycobiliprotein HaPE555 from the cryptophyte Hemiselmis andersenii CCMP644 in a tight interface filament | | Descriptor: | DiCys-(15,16)-Dihydrobiliverdin, PHYCOERYTHROBILIN, Phycoerythrin alpha-1 subunit, ... | | Authors: | Rathbone, H.W, Michie, K.A, Laos, A.L, Curmi, P.M.G. | | Deposit date: | 2022-09-23 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Molecular dissection of the soluble photosynthetic antenna from the cryptophyte alga Hemiselmis andersenii.
Commun Biol, 6, 2023
|
|
5LCC
 
 | | Oceanobacillus iheyensis macrodomain mutant D40A | | Descriptor: | MACROD-TYPE MACRODOMAIN | | Authors: | Gil-Ortiz, F, Zapata-Perez, R, Martinez, A.B, Juanhuix, J, Sanchez-Ferrer, A. | | Deposit date: | 2016-06-20 | | Release date: | 2017-05-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and functional analysis of Oceanobacillus iheyensis macrodomain reveals a network of waters involved in substrate binding and catalysis.
Open Biol, 7, 2017
|
|
8EHZ
 
 | | Crystal structure of the STUB1 TPR domain in complex with H317, a Helicon Polypeptide | | Descriptor: | E3 ubiquitin-protein ligase CHIP, H317, N,N'-(1,4-phenylene)diacetamide | | Authors: | Li, K, Swiecicki, J.-M, Tokareva, O.S, Thomson, T.M, Verdine, G.L, McGee, J.H. | | Deposit date: | 2022-09-14 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Recognition and reprogramming of E3 ubiquitin ligase surfaces by alpha-helical peptides.
Nat Commun, 14, 2023
|
|
8EI0
 
 | | Crystal structure of the STUB1 TPR domain in complex with H318, a Helicon Polypeptide | | Descriptor: | 1,2-ETHANEDIOL, E3 ubiquitin-protein ligase CHIP, H318, ... | | Authors: | Li, K, Swiecicki, J.-M, Tokareva, O.S, Thomson, T.M, Verdine, G.L, McGee, J.H. | | Deposit date: | 2022-09-14 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Recognition and reprogramming of E3 ubiquitin ligase surfaces by alpha-helical peptides.
Nat Commun, 14, 2023
|
|
8OXG
 
 | | Crystal structure of human methionine aminopeptidase-2 complexed with (3R,4S,5S,6R)-5-methoxy-4-[(2R,3R)-2-methyl-3-(3-methyl-2-buten-1-yl)-2-oxiranyl]-1-oxaspiro[2.5]oct-6-yl N-(trans-4-aminocyclohexyl)carbamate | | Descriptor: | MANGANESE (II) ION, Methionine aminopeptidase 2, [(1~{R},2~{S},3~{S},4~{R})-2-methoxy-4-methyl-3-[(2~{R},3~{S})-2-methyl-3-(3-methylbut-2-enyl)oxiran-2-yl]-4-oxidanyl-cyclohexyl] ~{N}-(4-azanylcyclohexyl)carbamate | | Authors: | Moss, S, Cornelius, P. | | Deposit date: | 2023-05-02 | | Release date: | 2024-04-10 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.731 Å) | | Cite: | Pharmacological Characterization of SDX-7320/Evexomostat: A Novel Methionine Aminopeptidase Type 2 Inhibitor with Anti-tumor and Anti-metastatic Activity.
Mol.Cancer Ther., 23, 2024
|
|
8EI3
 
 | | Crystal structure of VHL in complex with H313, a Helicon Polypeptide | | Descriptor: | Elongin-B, Elongin-C, H313, ... | | Authors: | Li, K, Tokareva, O.S, Thomson, T.M, Verdine, G.L, McGee, J.H. | | Deposit date: | 2022-09-14 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.49 Å) | | Cite: | Recognition and reprogramming of E3 ubiquitin ligase surfaces by alpha-helical peptides.
Nat Commun, 14, 2023
|
|
8EI6
 
 | | Crystal structure of the WWP2 HECT domain in complex with H305, a Helicon Polypeptide | | Descriptor: | H305, N,N'-(1,4-phenylene)diacetamide, NEDD4-like E3 ubiquitin-protein ligase WWP2 | | Authors: | Li, K, Tokareva, O.S, Thomson, T.M, Verdine, G.L, McGee, J.H. | | Deposit date: | 2022-09-14 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.62 Å) | | Cite: | Recognition and reprogramming of E3 ubiquitin ligase surfaces by alpha-helical peptides.
Nat Commun, 14, 2023
|
|
5M1O
 
 | | Crystal structure of the large terminase nuclease from thermophilic phage G20c with bound Cobalt | | Descriptor: | COBALT (II) ION, Phage terminase large subunit | | Authors: | Xu, R.G, Jenkins, H.T, Chechik, M, Blagova, E.V, Greive, S.J, Antson, A.A. | | Deposit date: | 2016-10-09 | | Release date: | 2016-10-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Viral genome packaging terminase cleaves DNA using the canonical RuvC-like two-metal catalysis mechanism.
Nucleic Acids Res., 45, 2017
|
|
8EI5
 
 | | Crystal structure of the WWP2 HECT domain in complex with H301, a Helicon Polypeptide | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, H301, ... | | Authors: | Li, K, Tokareva, O.S, Thomson, T.M, Verdine, G.L, McGee, J.H. | | Deposit date: | 2022-09-14 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Recognition and reprogramming of E3 ubiquitin ligase surfaces by alpha-helical peptides.
Nat Commun, 14, 2023
|
|
8EU1
 
 | |
5LIV
 
 | | Crystal structure of myxobacterial CYP260A1 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Cytochrome P450 CYP260A1,Cytochrome P450 CYP260A1, DIMETHYL SULFOXIDE, ... | | Authors: | Carius, Y, Khatri, Y, Bernhardt, R, Lancaster, C.R.D. | | Deposit date: | 2016-07-15 | | Release date: | 2016-11-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Structural characterization of CYP260A1 from Sorangium cellulosum to investigate the 1 alpha-hydroxylation of a mineralocorticoid.
FEBS Lett., 590, 2016
|
|
