5GHM
 
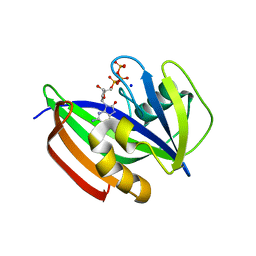 | | Crystal structure of human MTH1(G2K/D120N mutant) in complex with 8-oxo-dGTP at pH 7.0 | | Descriptor: | 7,8-dihydro-8-oxoguanine triphosphatase, 8-OXO-2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, SODIUM ION | | Authors: | Nakamura, T, Waz, S, Hirata, K, Nakabeppu, Y, Yamagata, Y. | | Deposit date: | 2016-06-20 | | Release date: | 2017-01-04 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural and Kinetic Studies of the Human Nudix Hydrolase MTH1 Reveal the Mechanism for Its Broad Substrate Specificity
J. Biol. Chem., 292, 2017
|
|
5G56
 
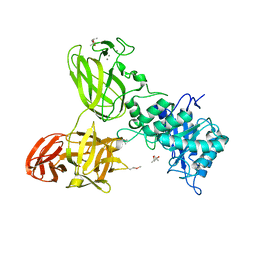 | | THE TETRA-MODULAR CELLULOSOMAL ARABINOXYLANASE CtXyl5A STRUCTURE AS REVEALED BY X-RAY CRYSTALLOGRAPHY | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, CARBOHYDRATE BINDING FAMILY 6 | | Authors: | Bras, J.L.A, Gilbert, H.J, Ferreira, L.M.A, Fontes, C.M.G.A, Najmudin, S. | | Deposit date: | 2016-05-21 | | Release date: | 2016-06-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | The Mechanism by which Arabinoxylanases Can Recognise Highly Decorated Xylans.
J.Biol.Chem., 291, 2016
|
|
5GSX
 
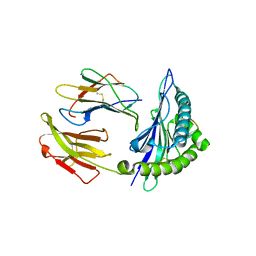 | | Mouse MHC class I H-2Kd with a MERS-CoV-derived peptide 142-2 | | Descriptor: | 10-mer peptide from Spike protein, Beta-2-microglobulin, H-2 class I histocompatibility antigen, ... | | Authors: | Liu, K, Chai, Y, Qi, J, Tan, W, Liu, W.J, Gao, G.F. | | Deposit date: | 2016-08-17 | | Release date: | 2017-06-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Protective T Cell Responses Featured by Concordant Recognition of Middle East Respiratory Syndrome Coronavirus-Derived CD8+ T Cell Epitopes and Host MHC.
J. Immunol., 198, 2017
|
|
5GXQ
 
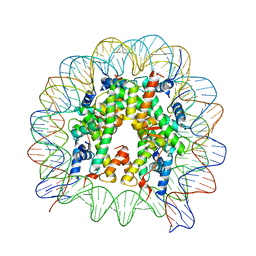 | | The crystal structure of the nucleosome containing H3.6 | | Descriptor: | DNA (146-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Taguchi, H, Xie, Y, Horikoshi, N, Kurumizaka, H. | | Deposit date: | 2016-09-19 | | Release date: | 2017-04-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystal Structure and Characterization of Novel Human Histone H3 Variants, H3.6, H3.7, and H3.8
Biochemistry, 56, 2017
|
|
2AYK
 
 | |
3R1G
 
 | | Structure Basis of Allosteric Inhibition of BACE1 by an Exosite-Binding Antibody | | Descriptor: | Beta-secretase 1, FAB of YW412.8.31 antibody heavy chain, FAB of YW412.8.31 antibody light chain | | Authors: | Wang, W, Rouge, L, Wu, P, Chiu, C, Chen, Y, Wu, Y, Watts, R.J. | | Deposit date: | 2011-03-10 | | Release date: | 2011-06-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A Therapeutic Antibody Targeting BACE1 Inhibits Amyloid-{beta} Production in Vivo.
Sci Transl Med, 3, 2011
|
|
8OMM
 
 | | Coproporphyrin III - LmCpfC complex soaked 3min with Fe2+ | | Descriptor: | 1,2-ETHANEDIOL, Coproporphyrin III ferrochelatase, FE (II) ION, ... | | Authors: | Gabler, T, Hofbauer, S. | | Deposit date: | 2023-03-31 | | Release date: | 2023-05-03 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Iron insertion into coproporphyrin III-ferrochelatase complex: Evidence for an intermediate distorted catalytic species.
Protein Sci., 32, 2023
|
|
8ONV
 
 | | KRAS-G13D in complex with BI-2493 | | Descriptor: | (7~{S})-2'-azanyl-3-[2-[(2~{S})-2-methylpiperazin-1-yl]pyrimidin-4-yl]spiro[5,6-dihydro-4~{H}-1,2-benzoxazole-7,4'-6,7-dihydro-5~{H}-1-benzothiophene]-3'-carbonitrile, 1,2-ETHANEDIOL, GTPase KRas, ... | | Authors: | Boettcher, J, Herdeis, L. | | Deposit date: | 2023-04-04 | | Release date: | 2023-06-07 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.01 Å) | | Cite: | Pan-KRAS inhibitor disables oncogenic signalling and tumour growth.
Nature, 619, 2023
|
|
2CRT
 
 | |
3CAV
 
 | | Crystal structure of 5beta-reductase (AKR1D1) in complex with NADP+ and 5beta-pregnan-3,20-dione | | Descriptor: | (5BETA)-PREGNANE-3,20-DIONE, 1,2-ETHANEDIOL, 3-oxo-5-beta-steroid 4-dehydrogenase, ... | | Authors: | Faucher, F, Cantin, L, Breton, R. | | Deposit date: | 2008-02-20 | | Release date: | 2008-07-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structure of human Delta4-3-ketosteroid 5beta-reductase defines the functional role of the residues of the catalytic tetrad in the steroid double bond reduction mechanism.
Biochemistry, 47, 2008
|
|
3POR
 
 | |
3TRA
 
 | |
4QOZ
 
 | |
2F1N
 
 | |
5GHQ
 
 | | Crystal structure of human MTH1(G2K/D120A mutant) in complex with 2-oxo-dATP under high concentrations of 2-oxo-dATP | | Descriptor: | 7,8-dihydro-8-oxoguanine triphosphatase, SODIUM ION, [[(2R,3S,5R)-5-(6-azanyl-2-oxidanylidene-1H-purin-9-yl)-3-oxidanyl-oxolan-2-yl]methoxy-oxidanyl-phosphoryl] phosphono hydrogen phosphate | | Authors: | Nakamura, T, Waz, S, Hirata, K, Nakabeppu, Y, Yamagata, Y. | | Deposit date: | 2016-06-20 | | Release date: | 2017-01-04 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.181 Å) | | Cite: | Structural and Kinetic Studies of the Human Nudix Hydrolase MTH1 Reveal the Mechanism for Its Broad Substrate Specificity
J. Biol. Chem., 292, 2017
|
|
5HHH
 
 | | Structure of human DNA polymerase beta Host-Guest complexed with the control G for N7-CBZ-platination | | Descriptor: | DNA (5'-D(*CP*CP*GP*AP*CP*GP*GP*AP*GP*GP*AP*GP*CP*AP*GP*G)-3'), DNA (5'-D(P*CP*CP*TP*GP*CP*TP*CP*CP*TP*C)-3'), DNA (5'-D(P*GP*TP*CP*GP*G)-3'), ... | | Authors: | Koag, M.-C, Lee, S. | | Deposit date: | 2016-01-11 | | Release date: | 2017-01-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.363 Å) | | Cite: | Synthesis, structure, and biological evaluation of a platinum-carbazole conjugate.
Chem Biol Drug Des, 91, 2018
|
|
5HHN
 
 | | Crystal Structure of HLA-A*0201 in complex with M1-F5L | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A-2 alpha chain, ... | | Authors: | Gras, S, Josephs, T.M, Rossjohn, J. | | Deposit date: | 2016-01-11 | | Release date: | 2016-03-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Molecular basis for universal HLA-A*0201-restricted CD8+ T-cell immunity against influenza viruses.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
3CH5
 
 | | The crystal structure of the RanGDP-Nup153ZnF2 complex | | Descriptor: | Fragment of Nuclear pore complex protein Nup153, GTP-binding nuclear protein Ran, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Vetter, I.R, Schrader, N. | | Deposit date: | 2008-03-07 | | Release date: | 2008-07-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Crystal Structure of the Ran-Nup153ZnF2 Complex: a General Ran Docking Site at the Nuclear Pore Complex
Structure, 16, 2008
|
|
6H58
 
 | | Structure of a hibernating 100S ribosome reveals an inactive conformation of the ribosomal protein S1 - Full 100S Hibernating E. coli Ribosome | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S1, ... | | Authors: | Beckert, B, Turk, M, Czech, A, Berninghausen, O, Beckmann, R, Ignatova, Z, Plitzko, J, Wilson, D.N. | | Deposit date: | 2018-07-24 | | Release date: | 2018-09-05 | | Last modified: | 2018-10-24 | | Method: | ELECTRON MICROSCOPY (7.9 Å) | | Cite: | Structure of a hibernating 100S ribosome reveals an inactive conformation of the ribosomal protein S1.
Nat Microbiol, 3, 2018
|
|
5HRD
 
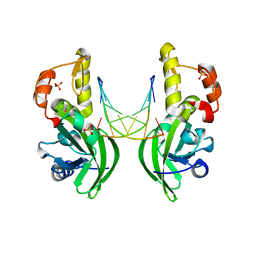 | |
5HRK
 
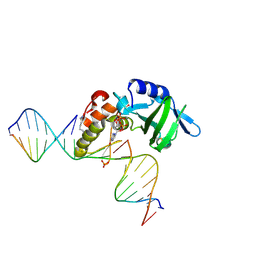 | |
5HXN
 
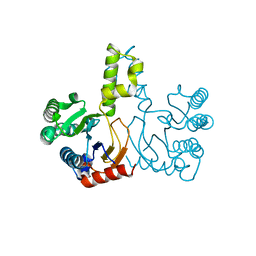 | | Crystal Structure of Z,Z-Farnesyl Diphosphate Synthase (D71M and E75A mutants) from the Wild Tomato Solanum habrochaites | | Descriptor: | (2Z,6Z)-farnesyl diphosphate synthase, chloroplastic | | Authors: | Lee, C.C, Chan, Y.T, Wang, A.H.J. | | Deposit date: | 2016-01-31 | | Release date: | 2017-04-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal Structure and Potential Head-to-Middle Condensation Function of aZ,Z-Farnesyl Diphosphate Synthase.
Acs Omega, 2, 2017
|
|
4JOO
 
 | | Spirocyclic Beta-Site Amyloid Precursor Protein Cleaving Enzyme 1 (BACE1) Inhibitors | | Descriptor: | (4R)-2'-amino-6-bromo-1',2,2-trimethyl-2,3-dihydrospiro[chromene-4,4'-imidazol]-5'(1'H)-one, Beta-secretase 1, NICKEL (II) ION | | Authors: | Vigers, G.P.A, Smith, D. | | Deposit date: | 2013-03-18 | | Release date: | 2013-04-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Spirocyclic beta-site amyloid precursor protein cleaving enzyme 1 (BACE1) inhibitors: from hit to lowering of cerebrospinal fluid (CSF) amyloid beta in a higher species.
J.Med.Chem., 56, 2013
|
|
3GSB
 
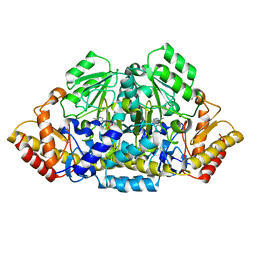 | |
2XTT
 
 | | Bovine trypsin in complex with evolutionary enhanced Schistocerca gregaria protease inhibitor 1 (SGPI-1-P02) | | Descriptor: | ACETATE ION, CALCIUM ION, CATIONIC TRYPSIN, ... | | Authors: | Wahlgren, W.Y, Pal, G, Kardos, J, Porrogi, P, Szenthe, B, Patthy, A, Graf, L, Katona, G. | | Deposit date: | 2010-10-12 | | Release date: | 2010-11-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (0.93 Å) | | Cite: | The catalytic aspartate is protonated in the Michaelis complex formed between trypsin and an in vitro evolved substrate-like inhibitor: a refined mechanism of serine protease action.
J.Biol.Chem., 286, 2011
|
|
