3CDM
 
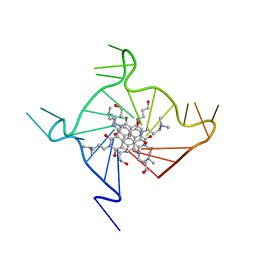 | | Structural adaptation and conservation in quadruplex-drug recognition | | Descriptor: | 2,7-bis[3-(dimethylamino)propyl]-4,9-bis[(3-hydroxypropyl)amino]benzo[lmn][3,8]phenanthroline-1,3,6,8(2H,7H)-tetrone, DNA (5'-D(*DT*DAP*DGP*DGP*DGP*DTP*DTP*DAP*DGP*DGP*DGP*DTP*DTP*DAP*DGP*DGP*DGP*DTP*DTP*DAP*DGP*DGP*DG)-3'), POTASSIUM ION | | Authors: | Parkinson, G.N, Neidle, S. | | Deposit date: | 2008-02-27 | | Release date: | 2008-09-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Topology conservation and loop flexibility in quadruplex-drug recognition: crystal structures of inter- and intramolecular telomeric DNA quadruplex-drug complexes
J.Mol.Biol., 381, 2008
|
|
6XA1
 
 | |
2WME
 
 | | Crystallographic structure of betaine aldehyde dehydrogenase from Pseudomonas aeruginosa | | Descriptor: | BETA-MERCAPTOETHANOL, BETAINE ALDEHYDE DEHYDROGENASE, GLYCEROL, ... | | Authors: | Gonzalez-Segura, L, Rudino-Pinera, E, Munoz-Clares, R.A, Horjales, E. | | Deposit date: | 2009-06-30 | | Release date: | 2009-08-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Crystal Structure of a Ternary Complex of Betaine Aldehyde Dehydrogenase from Pseudomonas Aeruginosa Provides New Insight Into the Reaction Mechanism and Shows a Novel Binding Mode of the 2'- Phosphate of Nadp(+) and a Novel Cation Binding Site.
J.Mol.Biol., 385, 2009
|
|
2XDR
 
 | | CRYSTALLOGRAPHIC STRUCTURE OF BETAINE ALDEHYDE DEHYDROGENASE MUTANT E252A FROM PSEUDOMONAS AERUGINOSA | | Descriptor: | 2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXYL, BETAINE ALDEHYDE DEHYDROGENASE, GLYCEROL, ... | | Authors: | Diaz-Sanchez, A.G, Gonzalez-Segura, L, Rudino-Pinera, E, Lira-Rocha, A, Munoz-Clares, R.A. | | Deposit date: | 2010-05-06 | | Release date: | 2011-06-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | A Novel Cysteine-Nadph Covalent Adduct in Pseudomonas Aeruginosa Betaine Aldehyde Dehydrogenase Suggests Important Roles for the Reduced Nucleotide in the Reaction Mechanism
To be Published
|
|
1MAG
 
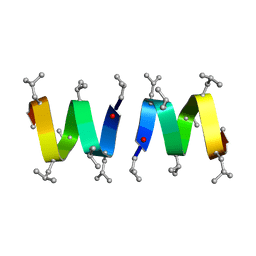 | |
3ZQA
 
 | | CRYSTALLOGRAPHIC STRUCTURE OF BETAINE ALDEHYDE DEHYDROGENASE MUTANT C286A FROM PSEUDOMONAS AERUGINOSA IN COMPLEX WITH NADPH | | Descriptor: | 1,2-ETHANEDIOL, 2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXYL, BETAINE ALDEHYDE DEHYDROGENASE, ... | | Authors: | Diaz-Sanchez, A.G, Gonzalez-Segura, L, Rudino-Pinera, E, Lira-Rocha, A, Torres-Larios, A, Munoz-Clares, R.A. | | Deposit date: | 2011-06-08 | | Release date: | 2011-10-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Novel Nadph-Cysteine Covalent Adduct Found in the Active Site of an Aldehyde Dehydrogenase.
Biochem.J., 439, 2011
|
|
4A0M
 
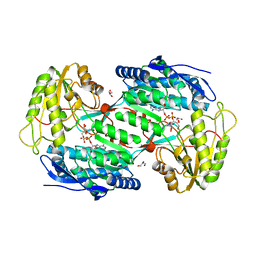 | | CRYSTAL STRUCTURE OF BETAINE ALDEHYDE DEHYDROGENASE FROM SPINACH IN COMPLEX WITH NAD | | Descriptor: | BETAINE ALDEHYDE DEHYDROGENASE, CHLOROPLASTIC, GLYCEROL, ... | | Authors: | Gonzalez-Segura, L, Rudino-Pinera, E, Diaz-Sanchez, A.G, Munoz-Clares, R.A. | | Deposit date: | 2011-09-09 | | Release date: | 2012-04-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Amino Acid Residues Critical for the Specificity for Betaine Aldehyde of the Plant Aldh10 Isoenzyme Involved in the Synthesis of Glycine Betaine.
Plant Physiol., 158, 2012
|
|
8G61
 
 | | mRNA decoding in human is kinetically and structurally distinct from bacteria (AC state) | | Descriptor: | 1,4-DIAMINOBUTANE, 18S rRNA, 28S rRNA, ... | | Authors: | Holm, M, Natchiar, K.S, Rundlet, E.J, Myasnikov, A.G, Altman, R.B, Blanchard, S.C. | | Deposit date: | 2023-02-14 | | Release date: | 2023-04-19 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | mRNA decoding in human is kinetically and structurally distinct from bacteria.
Nature, 617, 2023
|
|
3LKI
 
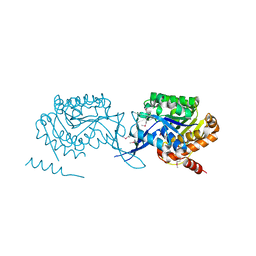 | |
8GLP
 
 | | mRNA decoding in human is kinetically and structurally distinct from bacteria (Consensus LSU focused refined structure) | | Descriptor: | 1,4-DIAMINOBUTANE, 18S rRNA, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Holm, M, Natchiar, K.S, Rundlet, E.J, Myasnikov, A.G, Watson, Z.L, Altman, R.B, Blanchard, S.C. | | Deposit date: | 2023-03-22 | | Release date: | 2023-04-19 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (1.67 Å) | | Cite: | mRNA decoding in human is kinetically and structurally distinct from bacteria.
Nature, 617, 2023
|
|
8G5Z
 
 | | mRNA decoding in human is kinetically and structurally distinct from bacteria (GA state) | | Descriptor: | 1,4-DIAMINOBUTANE, 18S rRNA, 28S rRNA, ... | | Authors: | Holm, M, Natchiar, K.S, Rundlet, E.J, Myasnikov, A.G, Altman, R.B, Blanchard, S.C. | | Deposit date: | 2023-02-14 | | Release date: | 2023-04-19 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.64 Å) | | Cite: | mRNA decoding in human is kinetically and structurally distinct from bacteria.
Nature, 617, 2023
|
|
8G6J
 
 | | mRNA decoding in human is kinetically and structurally distinct from bacteria (GA state 2) | | Descriptor: | (3R,6R,9S,12S,15S,18S,20R,24aR)-6-[(2S)-butan-2-yl]-3,12-bis[(1R)-1-hydroxy-2-methylpropyl]-8,9,11,17,18-pentamethyl-15-[(2S)-2-methylbutyl]hexadecahydropyrido[1,2-a][1,4,7,10,13,16,19]heptaazacyclohenicosine-1,4,7,10,13,16,19(21H)-heptone, (3beta)-O~3~-[(2R)-2,6-dihydroxy-2-(2-methoxy-2-oxoethyl)-6-methylheptanoyl]cephalotaxine, 1,4-DIAMINOBUTANE, ... | | Authors: | Holm, M, Natchiar, K.S, Rundlet, E.J, Myasnikov, A.G, Altman, R.B, Blanchard, S.C. | | Deposit date: | 2023-02-15 | | Release date: | 2023-04-19 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | mRNA decoding in human is kinetically and structurally distinct from bacteria.
Nature, 617, 2023
|
|
8G5Y
 
 | | mRNA decoding in human is kinetically and structurally distinct from bacteria (IC state) | | Descriptor: | 1,4-DIAMINOBUTANE, 18S rRNA, 28S rRNA, ... | | Authors: | Holm, M, Natchiar, K.S, Rundlet, E.J, Myasnikov, A.G, Altman, R.B, Blanchard, S.C. | | Deposit date: | 2023-02-14 | | Release date: | 2023-04-19 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.29 Å) | | Cite: | mRNA decoding in human is kinetically and structurally distinct from bacteria.
Nature, 617, 2023
|
|
8G60
 
 | | mRNA decoding in human is kinetically and structurally distinct from bacteria (CR state) | | Descriptor: | 1,4-DIAMINOBUTANE, 18S rRNA, 28S rRNA, ... | | Authors: | Holm, M, Natchiar, K.S, Rundlet, E.J, Myasnikov, A.G, Altman, R.B, Blanchard, S.C. | | Deposit date: | 2023-02-14 | | Release date: | 2023-04-19 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.54 Å) | | Cite: | mRNA decoding in human is kinetically and structurally distinct from bacteria.
Nature, 617, 2023
|
|
4IN9
 
 | | Structure of karilysin MMP-like catalytic domain in complex with inhibitory tetrapeptide SWFP | | Descriptor: | GLYCEROL, Karilysin protease, POTASSIUM ION, ... | | Authors: | Guevara, T, Ksiazek, M, Skottrup, P.D, Cerda-Costa, N, Trillo-Muyo, S, de Diego, I, Riise, E, Potempa, J, Gomis-Ruth, F.X. | | Deposit date: | 2013-01-04 | | Release date: | 2013-05-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure of the catalytic domain of the Tannerella forsythia matrix metallopeptidase karilysin in complex with a tetrapeptidic inhibitor.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
6F29
 
 | | Crystal structure of the kainate receptor GluK3 ligand binding domain in complex with (S)-1-[2-Amino-2-carboxyethyl]-5,7-dihydrothieno[3,4-d]pyrimidin-2,4(1H,3H)-dione at resolution 2.6A | | Descriptor: | (2~{S})-2-azanyl-3-[2,4-bis(oxidanylidene)-5,7-dihydrothieno[3,4-d]pyrimidin-1-yl]propanoic acid, CHLORIDE ION, Glutamate receptor ionotropic, ... | | Authors: | Venskutonyte, R, Frydenvang, K, Kastrup, J.S. | | Deposit date: | 2017-11-23 | | Release date: | 2018-02-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | ( S)-2-Amino-3-(5-methyl-3-hydroxyisoxazol-4-yl)propanoic Acid (AMPA) and Kainate Receptor Ligands: Further Exploration of Bioisosteric Replacements and Structural and Biological Investigation.
J. Med. Chem., 61, 2018
|
|
4EHQ
 
 | | Crystal Structure of Calmodulin Binding Domain of Orai1 in Complex with Ca2+/Calmodulin Displays a Unique Binding Mode | | Descriptor: | CALCIUM ION, Calcium release-activated calcium channel protein 1, Calmodulin, ... | | Authors: | Liu, Y, Zheng, X, Mueller, G.A, Sobhany, M, DeRose, E.F, Zhang, Y, London, R.E, Birnbaumer, L. | | Deposit date: | 2012-04-03 | | Release date: | 2012-11-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9005 Å) | | Cite: | Crystal structure of calmodulin binding domain of orai1 in complex with ca2+*calmodulin displays a unique binding mode.
J.Biol.Chem., 287, 2012
|
|
5JZX
 
 | | Crystal Structure of UDP-N-acetylenolpyruvoylglucosamine reductase (MurB) from Mycobacterium tuberculosis | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, POTASSIUM ION, UDP-N-acetylenolpyruvoylglucosamine reductase | | Authors: | Dharavath, S, Eniyan, K, Bajpai, U, Gourinath, S. | | Deposit date: | 2016-05-17 | | Release date: | 2017-05-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of UDP-N-acetylglucosamine-enolpyruvate reductase (MurB) from Mycobacterium tuberculosis
Biochim. Biophys. Acta, 1866, 2017
|
|
7M17
 
 | | SN-407-LRRC8A in MSP1E3D1 lipid nanodiscs (Pose-1) | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 7-{[(2S)-2-butyl-6,7-dichloro-2-cyclopentyl-1-oxo-2,3-dihydro-1H-inden-5-yl]oxy}heptanoic acid, Volume-regulated anion channel subunit LRRC8A | | Authors: | Kern, D.M, Gerber, E.E, Brohawn, S.G. | | Deposit date: | 2021-03-12 | | Release date: | 2021-03-24 | | Last modified: | 2022-02-23 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | Small molecule SWELL1 complex induction improves glycemic control and nonalcoholic fatty liver disease in murine Type 2 diabetes.
Nat Commun, 13, 2022
|
|
7M19
 
 | | SN-407-LRRC8A in MSP1E3D1 lipid nanodiscs (Pose-2) | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 7-{[(2S)-2-butyl-6,7-dichloro-2-cyclopentyl-1-oxo-2,3-dihydro-1H-inden-5-yl]oxy}heptanoic acid, Volume-regulated anion channel subunit LRRC8A | | Authors: | Kern, D.M, Gerber, E.E, Brohawn, S.G. | | Deposit date: | 2021-03-12 | | Release date: | 2021-03-24 | | Last modified: | 2022-02-23 | | Method: | ELECTRON MICROSCOPY (3.69 Å) | | Cite: | Small molecule SWELL1 complex induction improves glycemic control and nonalcoholic fatty liver disease in murine Type 2 diabetes.
Nat Commun, 13, 2022
|
|
3L8D
 
 | | Crystal structure of methyltransferase from Bacillus Thuringiensis | | Descriptor: | Methyltransferase, POTASSIUM ION | | Authors: | Fedorov, A.A, Fedorov, E.V, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-12-30 | | Release date: | 2010-01-12 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of methyltransferase from Bacillus Thuringiensis
To be Published
|
|
6T9R
 
 | | Aplysia californica AChBP in complex with a cytisine derivative | | Descriptor: | (1~{R},9~{S})-5-(3-oxidanylpropyl)-7,11-diazatricyclo[7.3.1.0^{2,7}]trideca-2,4-dien-6-one, 2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholine binding protein, ... | | Authors: | Davis, S, Hunter, W.N. | | Deposit date: | 2019-10-28 | | Release date: | 2020-02-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | The thermodynamic profile and molecular interactions of a C(9)-cytisine derivative-binding acetylcholine-binding protein from Aplysia californica.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
6RJN
 
 | | Crystal structure of a Fungal Catalase at 2.3 Angstroms | | Descriptor: | CHLORIDE ION, Catalase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Gomez, S, Navas-Yuste, S, Payne, A.M, Rivera, W, Lopez-Estepa, M, Brangbour, C, Fulla, D, Juanhuix, J, Fernandez, F.J, Vega, M.C. | | Deposit date: | 2019-04-28 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.295 Å) | | Cite: | Peroxisomal catalases from the yeasts Pichia pastoris and Kluyveromyces lactis as models for oxidative damage in higher eukaryotes.
Free Radic. Biol. Med., 141, 2019
|
|
2EZ2
 
 | | Apo tyrosine phenol-lyase from Citrobacter freundii at pH 8.0 | | Descriptor: | PHOSPHATE ION, POTASSIUM ION, Tyrosine phenol-lyase | | Authors: | Milic, D, Matkovic-Calogovic, D, Demidkina, T.V, Antson, A.A. | | Deposit date: | 2005-11-10 | | Release date: | 2006-07-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structures of apo- and holo-tyrosine phenol-lyase reveal a catalytically critical closed conformation and suggest a mechanism for activation by K+ ions
Biochemistry, 45, 2006
|
|
2G50
 
 | | The location of the allosteric amino acid binding site of muscle pyruvate kinase. | | Descriptor: | 1,2-ETHANEDIOL, 2-{2-[2-2-(METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, ALANINE, ... | | Authors: | Holyoak, T, Williams, R, Fenton, A.W. | | Deposit date: | 2006-02-22 | | Release date: | 2006-05-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Differentiating a Ligand's Chemical Requirements for Allosteric Interactions from Those for Protein Binding. Phenylalanine Inhibition of Pyruvate Kinase.
Biochemistry, 45, 2006
|
|
