1LQ8
 
 | | Crystal structure of cleaved protein C inhibitor | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Huntington, J.A, Kjellberg, M, Stenflo, J. | | Deposit date: | 2002-05-09 | | Release date: | 2003-02-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of Protein C Inhibitor Provides Insights into Hormone Binding and Heparin Activation
Structure, 11, 2003
|
|
2FM2
 
 | | HCV NS3-4A protease domain complexed with a ketoamide inhibitor, SCH446211 | | Descriptor: | BETA-MERCAPTOETHANOL, NS3 protease/helicase, NS4a protein, ... | | Authors: | Yi, M, Tong, X, Skelton, A, Chase, R, Chen, T, Prongay, A, Bogen, S.L, Saksena, A.K, Njoroge, F.G, Veselenak, R.L, Pyles, R.B, Bourne, N, Malcolm, B.A, Lemon, S.M. | | Deposit date: | 2006-01-06 | | Release date: | 2006-02-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Mutations conferring resistance to SCH6, a novel hepatitis C virus NS3/4A protease inhibitor. Reduced RNA replication fitness and partial rescue by second-site mutations
J.Biol.Chem., 281, 2006
|
|
1LKB
 
 | | Porcine Pancreatic Elastase/Na-Complex | | Descriptor: | CHLORIDE ION, Elastase 1, SODIUM ION, ... | | Authors: | Weiss, M.S, Panjikar, S, Nowak, E, Tucker, P.A. | | Deposit date: | 2002-04-24 | | Release date: | 2002-08-28 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Metal binding to porcine pancreatic elastase: calcium or not calcium.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
2FME
 
 | | Crystal structure of the mitotic kinesin eg5 (ksp) in complex with mg-adp and (r)-4-(3-hydroxyphenyl)-n,n,7,8-tetramethyl-3,4-dihydroisoquinoline-2(1h)-carboxamide | | Descriptor: | (4R)-4-(3-HYDROXYPHENYL)-N,N,7,8-TETRAMETHYL-3,4-DIHYDROISOQUINOLINE-2(1H)-CARBOXAMIDE, ADENOSINE-5'-DIPHOSPHATE, Kinesin-like protein KIF11, ... | | Authors: | Sheriff, S. | | Deposit date: | 2006-01-09 | | Release date: | 2006-04-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Inhibitors of human mitotic kinesin Eg5: Characterization of the 4-phenyl-tetrahydroisoquinoline lead series
Bioorg.Med.Chem.Lett., 16, 2006
|
|
1LAG
 
 | | STRUCTURAL BASES FOR MULTIPLE LIGAND SPECIFICITY OF THE PERIPLASMIC LYSINE-, ARGININE-, ORNITHINE-BINDING PROTEIN | | Descriptor: | HISTIDINE, LYSINE, ARGININE, ... | | Authors: | Kim, S.-H, Oh, B.-H. | | Deposit date: | 1993-10-06 | | Release date: | 1995-07-10 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structural basis for multiple ligand specificity of the periplasmic lysine-, arginine-, ornithine-binding protein.
J.Biol.Chem., 269, 1994
|
|
1LPG
 
 | | CRYSTAL STRUCTURE OF FXA IN COMPLEX WITH 79. | | Descriptor: | Blood coagulation factor Xa, CALCIUM ION, [4-({[5-BENZYLOXY-1-(3-CARBAMIMIDOYL-BENZYL)-1H-INDOLE-2-CARBONYL]-AMINO}-METHYL)-PHENYL]-TRIMETHYL-AMMONIUM | | Authors: | Schreuder, H.A, Brachvogel, V, Liesum, A. | | Deposit date: | 2002-05-08 | | Release date: | 2003-05-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design and Quantitative Structure-Activity relationship of 3-amidinobenzyl-1H-indole-2-carboxamides as potent, nonchiral, and selective inhibitors of blood coagulation factor Xa.
J.Med.Chem., 45, 2002
|
|
1LPZ
 
 | | CRYSTAL STRUCTURE OF FXA IN COMPLEX WITH 41. | | Descriptor: | 1-(3-carbamimidoylbenzyl)-N-(3,5-dichlorobenzyl)-4-methyl-1H-indole-2-carboxamide, Blood coagulation factor Xa, CALCIUM ION | | Authors: | Schreuder, H.A, Brachvogel, V, Liesum, A. | | Deposit date: | 2002-05-08 | | Release date: | 2003-05-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Design and Quantitative Structure-Activity relationship of 3-amidinobenzyl-1H-indole-2-carboxamides as potent, nonchiral, and selective inhibitors of blood coagulation factor Xa
J.Biol.Chem., 45, 2002
|
|
1LR4
 
 | | Room Temperature Crystal Structure of the Apo-form of the catalytic subunit of protein kinase CK2 from Zea mays | | Descriptor: | BENZAMIDINE, Protein kinase CK2 | | Authors: | Niefind, K, Puetter, M, Guerra, B, Issinger, O.-G, Schomburg, D. | | Deposit date: | 2002-05-14 | | Release date: | 2002-05-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Inclining the purine base binding plane in protein kinase CK2 by exchanging the flanking side-chains generates a preference for ATP as a cosubstrate.
J.Mol.Biol., 347, 2005
|
|
1IXC
 
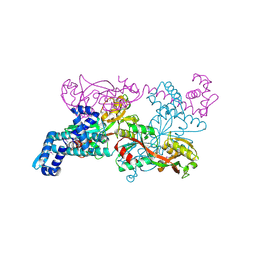 | | Crystal structure of CbnR, a LysR family transcriptional regulator | | Descriptor: | LysR-type regulatory protein | | Authors: | Muraoka, S, Okumura, R, Ogawa, N, Miyashita, K, Senda, T. | | Deposit date: | 2002-06-18 | | Release date: | 2003-06-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of a Full-length LysR-type Transcriptional Regulator, CbnR: Unusual Combination of Two Subunit Forms and Molecular Bases for Causing and Changing DNA Bend
J.Mol.Biol., 328, 2003
|
|
1IZH
 
 | | Inhibitor of HIV protease with unusual binding mode potently inhibiting multi-resistant protease mutants | | Descriptor: | proteinase, {(1S)-1-BENZYL-4-[3-CARBAMOYL-1-(1-CARBAMOYL-2-PHENYL-ETHYLCARBAMOYL)-(S)-PROPYLCARBAMOYL]-2-OXO-5-PHENYL-PENTYL}-CARBAMIC ACID TERT-BUTYL ESTER | | Authors: | Weber, J, Mesters, J.R, Lepsik, M, Prejdova, J, Svec, M, Sponarova, J, Mlcochova, P, Skalicka, K, Strisovsky, K, Uhlikova, T, Soucek, M, Machala, L, Stankova, M, Vondrasek, J, Klimkait, T, Kraeusslich, H.-G, Hilgenfeld, R, Konvalinka, J. | | Deposit date: | 2002-10-02 | | Release date: | 2002-12-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Unusual Binding Mode of an HIV-1 Protease Inhibitor Explains its Potency against Multi-drug-resistant Virus Strains
J.MOL.BIOL., 324, 2002
|
|
1IZ1
 
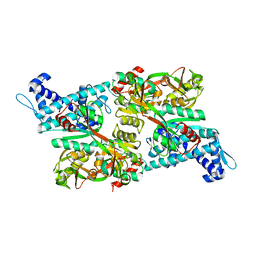 | | CRYSTAL STRUCTURE OF CBNR, A LYSR FAMILY TRANSCRIPTIONAL REGULATOR | | Descriptor: | LysR-type regulatory protein | | Authors: | Muraoka, S, Okumura, R, Ogawa, N, Miyashita, K, Senda, T. | | Deposit date: | 2002-09-18 | | Release date: | 2003-05-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of a Full-length LysR-type Transcriptional Regulator, CbnR: Unusual Combination of Two Subunit Forms and Molecular Bases for Causing and Changing DNA Bend
J.Mol.Biol., 328, 2003
|
|
1J2J
 
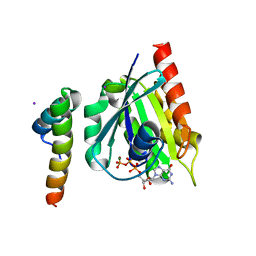 | | Crystal structure of GGA1 GAT N-terminal region in complex with ARF1 GTP form | | Descriptor: | ADP-ribosylation factor 1, ADP-ribosylation factor binding protein GGA1, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Shiba, T, Kawasaki, M, Takatsu, H, Nogi, T, Matsugaki, N, Igarashi, N, Suzuki, M, Kato, R, Nakayama, K, Wakatsuki, S. | | Deposit date: | 2003-01-05 | | Release date: | 2003-05-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Molecular mechanism of membrane recruitment of GGA by ARF in lysosomal protein transport
NAT.STRUCT.BIOL., 10, 2003
|
|
1J2U
 
 | | Creatininase Zn | | Descriptor: | SULFATE ION, ZINC ION, creatinine amidohydrolase | | Authors: | Yoshimoto, T, Tanaka, N, Kanada, N, Inoue, T, Nakajima, Y, Haratake, M, Nakamura, K.T, Xu, Y, Ito, K. | | Deposit date: | 2003-01-11 | | Release date: | 2004-01-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structures of creatininase reveal the substrate binding site and provide an insight into the catalytic mechanism
J.Mol.Biol., 337, 2004
|
|
1J54
 
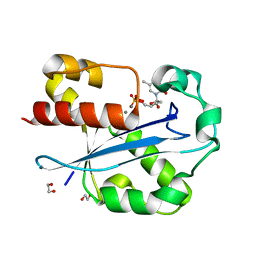 | | Structure of the N-terminal exonuclease domain of the epsilon subunit of E.coli DNA polymerase III at pH 5.8 | | Descriptor: | 1,2-ETHANEDIOL, DNA polymerase III, epsilon chain, ... | | Authors: | Hamdan, S, Carr, P.D, Brown, S.E, Ollis, D.L, Dixon, N.E. | | Deposit date: | 2002-01-22 | | Release date: | 2002-10-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis for Proofreading during Replication of the Escherichia coli Chromosome
Structure, 10, 2002
|
|
1IIQ
 
 | | CRYSTAL STRUCTURE OF HIV-1 PROTEASE COMPLEXED WITH A HYDROXYETHYLAMINE PEPTIDOMIMETIC INHIBITOR | | Descriptor: | GLYCEROL, N-{(2R,3S)-3-[(tert-butoxycarbonyl)amino]-2-hydroxy-4-phenylbutyl}-L-phenylalanyl-L-glutaminyl-L-phenylalaninamide, PROTEASE RETROPEPSIN | | Authors: | Dohnalek, J, Hasek, J, Duskova, J, Petrokova, H, Hradilek, M, Soucek, M, Konvalinka, J, Brynda, J, Sedlacek, J, Fabry, M. | | Deposit date: | 2001-04-24 | | Release date: | 2002-04-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Hydroxyethylamine isostere of an HIV-1 protease inhibitor prefers its amine to the hydroxy group in binding to catalytic aspartates. A synchrotron study of HIV-1 protease in complex with a peptidomimetic inhibitor.
J.Med.Chem., 45, 2002
|
|
2E99
 
 | | E. coli undecaprenyl pyrophosphate synthase in complex with BPH-608 | | Descriptor: | (1-HYDROXY-1-PHOSPHONO-2-[1,1';3',1'']TERPHENYL-3-YL-ETHYL)-PHOSPHONIC ACID, Undecaprenyl pyrophosphate synthetase | | Authors: | Guo, R.T, Ko, T.P, Cao, R, Liang, P.H, Oldfield, E, Wang, A.H.J. | | Deposit date: | 2007-01-24 | | Release date: | 2007-06-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Bisphosphonates target multiple sites in both cis- and trans-prenyltransferases
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2E8X
 
 | | S. cerevisiae geranylgeranyl pyrophosphate synthase in complex with magnesium and GPP | | Descriptor: | GERANYL DIPHOSPHATE, Geranylgeranyl pyrophosphate synthetase, MAGNESIUM ION | | Authors: | Guo, R.T, Ko, T.P, Chen, C.K.-M, Jeng, W.Y, Chang, T.H, Liang, P.H, Oldfield, E, Wang, A.H.-J. | | Deposit date: | 2007-01-24 | | Release date: | 2007-06-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Bisphosphonates target multiple sites in both cis- and trans-prenyltransferases
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2E93
 
 | | S. cerevisiae geranylgeranyl pyrophosphate synthase in complex with BPH-629 | | Descriptor: | Geranylgeranyl pyrophosphate synthetase, [2-(3-DIBENZOFURAN-4-YL-PHENYL)-1-HYDROXY-1-PHOSPHONO-ETHYL]-PHOSPHONIC ACID | | Authors: | Guo, R.T, Cao, R, Ko, T.P, Chen, C.K.-M, Jeng, W.Y, Chang, T.H, Liang, P.H, Oldfield, E, Wang, A.H.-J. | | Deposit date: | 2007-01-24 | | Release date: | 2007-06-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Bisphosphonates target multiple sites in both cis- and trans-prenyltransferases
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2EH7
 
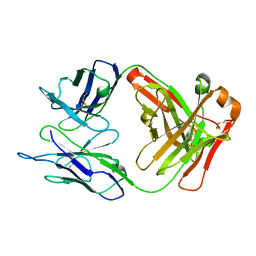 | | Crystal structure of humanized KR127 FAB | | Descriptor: | HUMANIZED KR127 FAB, HEAVY CHAIN, LIGHT CHAIN | | Authors: | Chi, S.-W, Kim, S.-J, Maeng, C.-Y, Hong, H.J, Ryu, S.-E. | | Deposit date: | 2007-03-05 | | Release date: | 2008-01-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Broadly neutralizing anti-hepatitis B virus antibody reveals a complementarity determining region H3 lid-opening mechanism
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
1IUQ
 
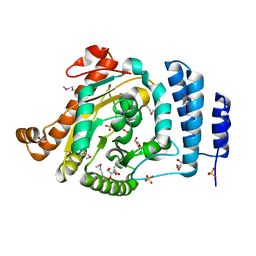 | | The 1.55 A Crystal Structure of Glycerol-3-Phosphate Acyltransferase | | Descriptor: | GLYCEROL, Glycerol-3-Phosphate Acyltransferase, SULFATE ION | | Authors: | Tamada, T, Feese, M.D, Kato, Y, Kuroki, R. | | Deposit date: | 2002-03-06 | | Release date: | 2003-10-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Substrate recognition and selectivity of plant glycerol-3-phosphate acyltransferases (GPATs) from Cucurbita moscata and Spinacea oleracea.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1IV7
 
 | |
1J3Y
 
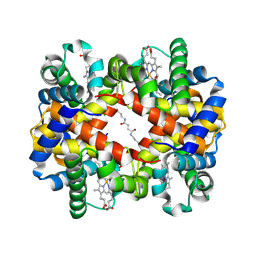 | | Direct observation of photolysis-induced tertiary structural changes in human hemoglobin; Crystal structure of alpha(Fe)-beta(Ni) hemoglobin (laser photolysed) | | Descriptor: | BUT-2-ENEDIAL, CARBON MONOXIDE, Hemoglobin alpha Chain, ... | | Authors: | Adachi, S, Park, S.-Y, Tame, J.R.H, Shiro, Y, Shibayama, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-02-21 | | Release date: | 2003-07-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Direct observation of photolysis-induced tertiary structural changes in hemoglobin
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1LQE
 
 | | CRYSTAL STRUCTURE OF TRYPSIN IN COMPLEX WITH 79. | | Descriptor: | CALCIUM ION, SULFATE ION, TRYPSIN, ... | | Authors: | Schreuder, H.A, Liesum, A. | | Deposit date: | 2002-05-10 | | Release date: | 2003-05-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Design and quantitative structure-activity relationship of 3-amidinobenzyl-1H-indole-2-carboxamides
as potent, nonchiral, and selective inhibitors of blood coagulation factor Xa.
J.Med.Chem., 45, 2002
|
|
1LR7
 
 | |
1KZ8
 
 | | CRYSTAL STRUCTURE OF PORCINE FRUCTOSE-1,6-BISPHOSPHATASE COMPLEXED WITH A NOVEL ALLOSTERIC-SITE INHIBITOR | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose, ADENOSINE MONOPHOSPHATE, FRUCTOSE-1,6-BISPHOSPHATASE, ... | | Authors: | Wright, S.W, Carlo, A.A, Carty, M.D, Danley, D.E, Hageman, D.L, Karam, G.A, Levy, C.B, Mansour, M.N, Mathiowetz, A.M, McClure, L.D, Nestor, N.B, McPherson, R.K, Pandit, J, Pustilnik, L.R, Schulte, G.K, Soeller, W.C, Treadway, J.L, Wang, I.-K, Bauer, P.H. | | Deposit date: | 2002-02-06 | | Release date: | 2002-10-16 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | ANILINOQUINAZOLINE INHIBITORS OF FRUCTOSE 1,6-BISPHOSPHATASE BIND AT A NOVEL ALLOSTERIC SITE: SYNTHESIS, IN VITRO CHARACTERIZATION, AND X-RAY CRYSTALLOGRAPHY
J.MED.CHEM., 45, 2002
|
|
