6X32
 
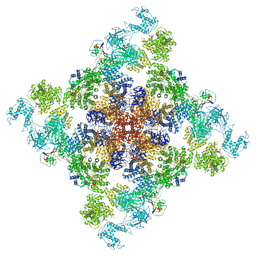 | | Wt pig RyR1 in complex with apoCaM, EGTA condition (class 1 and 2, closed) | | Descriptor: | Calmodulin-1, Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine Receptor, ... | | Authors: | Woll, K.W, Haji-Ghassemi, O, Van Petegem, F. | | Deposit date: | 2020-05-21 | | Release date: | 2021-01-13 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Pathological conformations of disease mutant Ryanodine Receptors revealed by cryo-EM.
Nat Commun, 12, 2021
|
|
7TZC
 
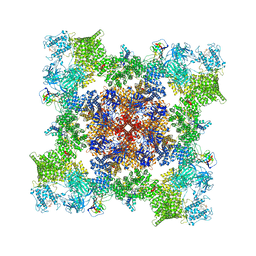 | | A drug and ATP binding site in type 1 ryanodine receptor | | Descriptor: | (2S)-3-(octadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 4-[(7-methoxy-2,3-dihydro-1,4-benzothiazepin-4(5H)-yl)methyl]benzoic acid, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Melville, Z, Dridi, H, Yuan, Q, Reiken, S, Anetta, W, Liu, Y, Clarke, O.B, Marks, A.R. | | Deposit date: | 2022-02-15 | | Release date: | 2022-05-18 | | Last modified: | 2022-07-20 | | Method: | ELECTRON MICROSCOPY (2.45 Å) | | Cite: | A drug and ATP binding site in type 1 ryanodine receptor.
Structure, 30, 2022
|
|
6EUS
 
 | |
2MW7
 
 | |
2ASC
 
 | | Scorpion toxin LQH-alpha-IT | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, ETHANOL, ... | | Authors: | Kahn, R, Karbat, I, Gurevitz, M, Frolow, F. | | Deposit date: | 2005-08-23 | | Release date: | 2006-09-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | X-ray structures of Lqh-alpha-IT and Lqh-alpha-IT8D9D10V mutant
To be Published
|
|
1SK6
 
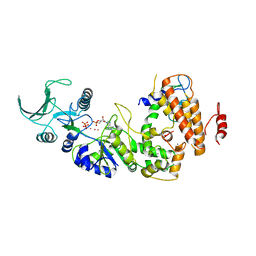 | | Crystal structure of the adenylyl cyclase domain of anthrax edema factor (EF) in complex with calmodulin, 3',5' cyclic AMP (cAMP), and pyrophosphate | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, CALCIUM ION, Calmodulin, ... | | Authors: | Guo, Q, Shen, Y, Zhukovskaya, N.L, Tang, W.J. | | Deposit date: | 2004-03-04 | | Release date: | 2004-06-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural and kinetic analyses of the interaction of anthrax adenylyl cyclase toxin with reaction products cAMP and pyrophosphate.
J.Biol.Chem., 279, 2004
|
|
7S1X
 
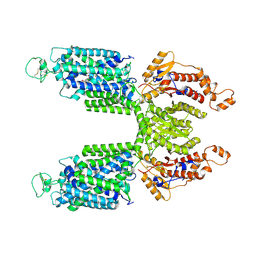 | |
7S1Y
 
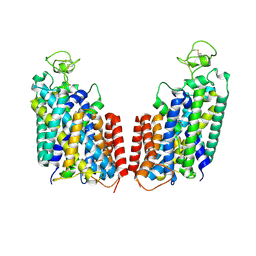 | |
5XRZ
 
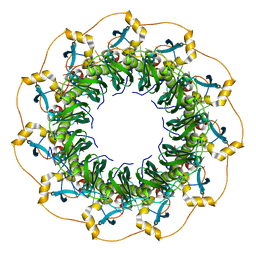 | | Structure of a ssDNA bound to the inner DNA binding site of RAD52 | | Descriptor: | DNA repair protein RAD52 homolog, POTASSIUM ION, ssDNA (40-MER) | | Authors: | Saotome, M, Saito, K, Yasuda, T, Sugiyama, S, Kurumizaka, H, Kagawa, W. | | Deposit date: | 2017-06-11 | | Release date: | 2018-04-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structural Basis of Homology-Directed DNA Repair Mediated by RAD52
iScience, 3, 2018
|
|
6M7H
 
 | | Structure of calmodulin with KN93 | | Descriptor: | CALCIUM ION, Calmodulin-1, N-[2-[[[3-(4'-Chlorophenyl)-2-propenyl]methylamino]methyl]phenyl]-N-(2-hydroxyethyl)-4'-methoxybenzenesulfonamide | | Authors: | Damo, S.M, Pattanayek, R, Johnson, C.N. | | Deposit date: | 2018-08-20 | | Release date: | 2019-08-28 | | Last modified: | 2019-11-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The CaMKII inhibitor KN93-calmodulin interaction and implications for calmodulin tuning of NaV1.5 and RyR2 function.
Cell Calcium, 82, 2019
|
|
7A56
 
 | | Schmallenberg Virus Envelope Glycoprotein Gc Fusion Domains in Postfusion Conformation | | Descriptor: | CHLORIDE ION, Envelopment polyprotein, PHOSPHATE ION, ... | | Authors: | Hellert, J, Guardado-Calvo, P, Rey, F.A. | | Deposit date: | 2020-08-20 | | Release date: | 2021-09-01 | | Last modified: | 2023-03-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure, function, and evolution of the Orthobunyavirus membrane fusion glycoprotein.
Cell Rep, 42, 2023
|
|
1M6V
 
 | | Crystal Structure of the G359F (small subunit) Point Mutant of Carbamoyl Phosphate Synthetase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, L-ornithine, ... | | Authors: | Thoden, J.B, Huang, X, Raushel, F.M, Holden, H.M. | | Deposit date: | 2002-07-17 | | Release date: | 2002-07-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Carbamoyl-phosphate synthetase. Creation of an escape route for ammonia
J.Biol.Chem., 277, 2002
|
|
6JIU
 
 | | Structure of RyR2 (F/A/C/L-Ca2+/Ca2+CaM dataset) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CAFFEINE, CALCIUM ION, ... | | Authors: | Gong, D.S, Chi, X.M, Zhou, G.W, Huang, G.X.Y, Lei, J.L, Yan, N. | | Deposit date: | 2019-02-23 | | Release date: | 2019-07-17 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Modulation of cardiac ryanodine receptor 2 by calmodulin.
Nature, 572, 2019
|
|
6JI8
 
 | | Structure of RyR2 (F/apoCaM dataset) | | Descriptor: | Calmodulin-1, Peptidyl-prolyl cis-trans isomerase FKBP1B, RyR2, ... | | Authors: | Gong, D.S, Chi, X.M, Zhou, G.W, Huang, G.X.Y, Lei, J.L, Yan, N. | | Deposit date: | 2019-02-20 | | Release date: | 2019-07-17 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Modulation of cardiac ryanodine receptor 2 by calmodulin.
Nature, 572, 2019
|
|
6JIY
 
 | | Structure of RyR2 (F/A/C/H-Ca2+/Ca2+CaM dataset) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CAFFEINE, CALCIUM ION, ... | | Authors: | Gong, D.S, Chi, X.M, Zhou, G.W, Huang, G.X.Y, Lei, J.L, Yan, N. | | Deposit date: | 2019-02-24 | | Release date: | 2019-07-17 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Modulation of cardiac ryanodine receptor 2 by calmodulin.
Nature, 572, 2019
|
|
6JRS
 
 | | Structure of RyR2 (*F/A/C/L-Ca2+/Ca2+-CaM dataset) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CAFFEINE, CALCIUM ION, ... | | Authors: | Gong, D.S, Chi, X.M, Zhou, G.W, Huang, G.X.Y, Lei, J.L, Yan, N. | | Deposit date: | 2019-04-05 | | Release date: | 2019-07-17 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Modulation of cardiac ryanodine receptor 2 by calmodulin.
Nature, 572, 2019
|
|
1R1G
 
 | |
6NW8
 
 | | SOLUTION STRUCTURE OF CN29, A TOXIN FROM CENTRUROIDES NOXIUS SCORPION VENOM | | Descriptor: | Cn29 | | Authors: | Delepierre, M, Gurrola, G.B, Possani, L.D, Guijarro, J.I. | | Deposit date: | 2019-02-06 | | Release date: | 2019-07-03 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Cn29, a novel orphan peptide found in the venom of the scorpion Centruroides noxius: Structure and function.
Toxicon, 167, 2019
|
|
6JII
 
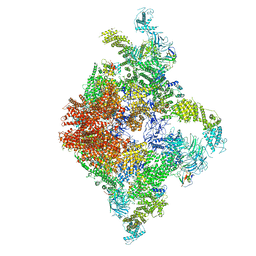 | | Structure of RyR2 (F/A/C/L-Ca2+/apo-CaM-M dataset) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CAFFEINE, CALCIUM ION, ... | | Authors: | Gong, D.S, Chi, X.M, Zhou, G.W, Huang, G.X.Y, Lei, J.L, Yan, N. | | Deposit date: | 2019-02-21 | | Release date: | 2019-07-17 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Modulation of cardiac ryanodine receptor 2 by calmodulin.
Nature, 572, 2019
|
|
2LO7
 
 | |
7KL5
 
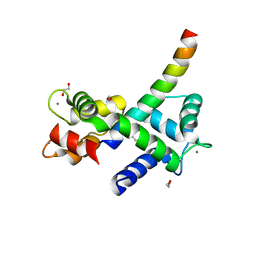 | | Structure of Calmodulin Bound to the Cardiac Ryanodine Receptor (RyR2) at Residues: Phe4246 to Val4271 | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Calmodulin-1, ... | | Authors: | Fisher, A.J, Ames, J.B, Yu, Q. | | Deposit date: | 2020-10-29 | | Release date: | 2021-04-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The Crystal Structure of Calmodulin Bound to the Cardiac Ryanodine Receptor (RyR2) at Residues Phe4246-Val4271 Reveals a Fifth Calcium Binding Site.
Biochemistry, 60, 2021
|
|
6JV2
 
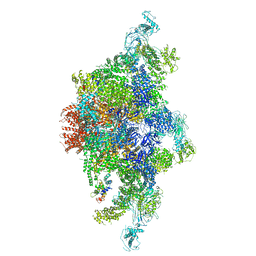 | | Structure of RyR2 (P/L-Ca2+/Ca2+-CaM dataset) | | Descriptor: | CALCIUM ION, Calmodulin-1, RyR2, ... | | Authors: | Gong, D.S, Chi, X.M, Zhou, G.W, Huang, G.X.Y, Lei, J.L, Yan, N. | | Deposit date: | 2019-04-15 | | Release date: | 2019-07-17 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Modulation of cardiac ryanodine receptor 2 by calmodulin.
Nature, 572, 2019
|
|
6RJR
 
 | | Crystal structure of a Fungal Catalase at 1.9 Angstrom | | Descriptor: | CHLORIDE ION, Catalase, GLYCEROL, ... | | Authors: | Gomez, S, Navas-Yuste, S, Payne, A.M, Rivera, W, Lopez-Estepa, M, Brangbour, C, Fulla, D, Juanhuix, J, Fernandez, F.J, Vega, M.C. | | Deposit date: | 2019-04-29 | | Release date: | 2020-03-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.895 Å) | | Cite: | Peroxisomal catalases from the yeasts Pichia pastoris and Kluyveromyces lactis as models for oxidative damage in higher eukaryotes.
Free Radic. Biol. Med., 141, 2019
|
|
6T9R
 
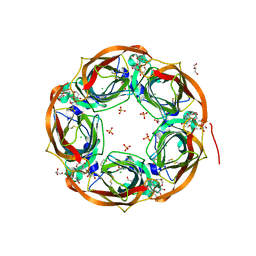 | | Aplysia californica AChBP in complex with a cytisine derivative | | Descriptor: | (1~{R},9~{S})-5-(3-oxidanylpropyl)-7,11-diazatricyclo[7.3.1.0^{2,7}]trideca-2,4-dien-6-one, 2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholine binding protein, ... | | Authors: | Davis, S, Hunter, W.N. | | Deposit date: | 2019-10-28 | | Release date: | 2020-02-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | The thermodynamic profile and molecular interactions of a C(9)-cytisine derivative-binding acetylcholine-binding protein from Aplysia californica.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
4H1T
 
 | | X-RAY Structure of the Complex VchUPh with Phosphate ion at 1.92A Resolution. | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Prokofev, I.I, Lashkov, A.A, Gabdoulkhakov, A.G, Sotnichenko, S.E, Betzel, C, Mikhailov, A.M. | | Deposit date: | 2012-09-11 | | Release date: | 2013-09-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.924 Å) | | Cite: | X-RAY Structure of the Complex VchUPh with Phosphate ion at 1.92A Resolution.
TO BE PUBLISHED
|
|
