4NJT
 
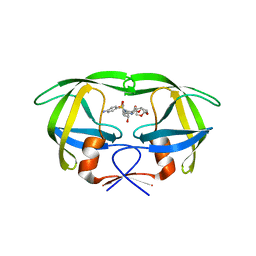 | | Crystal structure of multidrug-resistant clinical isolate A02 HIV-1 protease in complex with darunavir | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, Protease | | Authors: | Yedidi, R.S, Garimella, H, Chang, S.B, Kaufman, J.D, Das, D, Wingfield, P.T, Ghosh, A.K, Mitsuya, H. | | Deposit date: | 2013-11-11 | | Release date: | 2014-04-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A Conserved Hydrogen-Bonding Network of P2 bis-Tetrahydrofuran-Containing HIV-1 Protease Inhibitors (PIs) with a Protease Active-Site Amino Acid Backbone Aids in Their Activity against PI-Resistant HIV.
Antimicrob.Agents Chemother., 58, 2014
|
|
8XM4
 
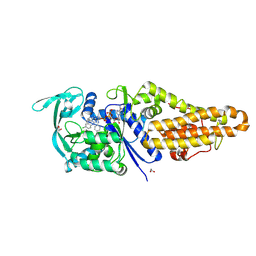 | | Methionyl-tRNA synthetase from Staphylococcus aureus in complex with chlorhexidine and ATP | | Descriptor: | 1,2-ETHANEDIOL, 1-[6-[azanylidene-[[azanylidene-[[(4-chlorophenyl)amino]methyl]-$l^{4}-azanyl]methyl]-$l^{4}-azanyl]hexyl]-3-[~{N}-(4-chlorophenyl)carbamimidoyl]guanidine, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Lu, F, Xia, K, Yi, J, Chen, B, Luo, Z, Xu, J, Gu, Q, Zhou, H. | | Deposit date: | 2023-12-27 | | Release date: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Biochemical and structural characterization of chlorhexidine as an ATP-assisted inhibitor against type 1 methionyl-tRNA synthetase from Gram-positive bacteria.
Eur.J.Med.Chem., 268, 2024
|
|
6HRV
 
 | |
5JQS
 
 | |
4NXS
 
 | | Crystal structure of human alpha-galactosidase A in complex with 1-deoxygalactonojirimycin-pFPhT | | Descriptor: | (2R,3S,4R,5S)-N-(4-fluorophenyl)-3,4,5-trihydroxy-2-(hydroxymethyl)piperidine-1-carbothioamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Johnson, J.L, Drury, J.E, Lieberman, R.L. | | Deposit date: | 2013-12-09 | | Release date: | 2014-06-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5493 Å) | | Cite: | Molecular Basis of 1-Deoxygalactonojirimycin Arylthiourea Binding to Human alpha-Galactosidase A: Pharmacological Chaperoning Efficacy on Fabry Disease Mutants.
Acs Chem.Biol., 9, 2014
|
|
7ZAU
 
 | | Fascin-1 in complex with Nb 3E11 | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Fascin, ... | | Authors: | Burgess, S.G, Bayliss, R. | | Deposit date: | 2022-03-22 | | Release date: | 2023-09-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A nanobody inhibitor of Fascin-1 actin-bundling activity and filopodia formation.
Open Biology, 14, 2024
|
|
3NCZ
 
 | |
5IPE
 
 | | Human Histidine Triad Nucleotide Binding Protein 1 (hHint1) nucleoside thiophosphoramidate catalytic product complex | | Descriptor: | 1,2-ETHANEDIOL, 5'-S-phosphono-5'-thioguanosine, CHLORIDE ION, ... | | Authors: | Maize, K.M, Finzel, B.C. | | Deposit date: | 2016-03-09 | | Release date: | 2017-03-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Caught before Released: Structural Mapping of the Reaction Trajectory for the Sofosbuvir Activating Enzyme, Human Histidine Triad Nucleotide Binding Protein 1 (hHint1).
Biochemistry, 56, 2017
|
|
4O44
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase in complex with 4-((4-(mesitylamino)-6-(3-morpholinopropoxy)-1,3,5-triazin-2-yl)amino)benzonitrile (JLJ529), a non-nucleoside inhibitor | | Descriptor: | 4-({4-[3-(morpholin-4-yl)propoxy]-6-[(2,4,6-trimethylphenyl)amino]-1,3,5-triazin-2-yl}amino)benzonitrile, HIV-1 reverse transcriptase, p51 subunit, ... | | Authors: | Mislak, A.C, Frey, K.M, Anderson, K.S. | | Deposit date: | 2013-12-18 | | Release date: | 2014-05-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.889 Å) | | Cite: | A mechanistic and structural investigation of modified derivatives of the diaryltriazine class of NNRTIs targeting HIV-1 reverse transcriptase.
Biochim.Biophys.Acta, 1840, 2014
|
|
5I9J
 
 | | Structure of the cholesterol and lutein-binding domain of human STARD3 at 1.74A | | Descriptor: | 1,2-ETHANEDIOL, L(+)-TARTARIC ACID, SULFATE ION, ... | | Authors: | Horvath, M.P, Bernstein, P.S, Li, B, George, E.W, Tran, Q.T. | | Deposit date: | 2016-02-20 | | Release date: | 2016-03-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structure of the lutein-binding domain of human StARD3 at 1.74 angstrom resolution and model of a complex with lutein.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
6RCG
 
 | | Crystal structure of Casein kinase 1 delta (CK1 delta) complexed with SR3029 inhibitor | | Descriptor: | 1,2-ETHANEDIOL, Casein kinase I isoform delta, ~{N}-[[6,7-bis(fluoranyl)-1~{H}-benzimidazol-2-yl]methyl]-9-(3-fluorophenyl)-2-morpholin-4-yl-purin-6-amine | | Authors: | Chaikuad, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Roush, W.R, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-04-11 | | Release date: | 2020-03-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of Casein kinase 1 delta (CK1 delta) complexed with SR3029 inhibitor
To Be Published
|
|
8I3P
 
 | | crystal structure of yeast cytosine deaminase mutant yCD-RQ-1/8SAH in complex with (R)-4-hydroxy-3,4-dihydropyrimidin-2(1H)-one | | Descriptor: | 1,2-ETHANEDIOL, 4-oxidanyl-3,4-dihydro-1H-pyrimidin-2-one, Cytosine deaminase, ... | | Authors: | Qin, M.M, Deng, H.Z, Yao, L.S. | | Deposit date: | 2023-01-17 | | Release date: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | crystal structure of yeast cytosine deaminase mutant yCD-RQ-1/8SAH in complex with (R)-4-hydroxy-3,4-dihydropyrimidin-2(1H)-one
To be published
|
|
8XOV
 
 | | The Crystal Structure of N-terminal kinase domain of human RSK-1 from Biortus. | | Descriptor: | 1,2-ETHANEDIOL, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Ribosomal protein S6 kinase alpha-1 | | Authors: | Wang, F, Cheng, W, Lv, Z, Meng, Q, Zhang, B. | | Deposit date: | 2024-01-02 | | Release date: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | The Crystal Structure of N-terminal kinase domain of human RSK-1 from Biortus.
To Be Published
|
|
8K69
 
 | | Cryo-EM structure of Oryza sativa HKT2;2/1 at 2.3 angstrom | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CHOLESTEROL, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Wang, X, Shen, X, Qu, Y, Wang, C, Shen, H. | | Deposit date: | 2023-07-25 | | Release date: | 2024-04-03 | | Last modified: | 2025-07-23 | | Method: | ELECTRON MICROSCOPY (2.33 Å) | | Cite: | Structural insights into ion selectivity and transport mechanisms of Oryza sativa HKT2;1 and HKT2;2/1 transporters.
Nat.Plants, 10, 2024
|
|
7UE1
 
 | | HIV-1 Integrase Catalytic Core Domain Mutant (KGD) in Complex with Inhibitor GRL-142 | | Descriptor: | (3S,3aR,5R,7aS,8S)-hexahydro-4H-3,5-methanofuro[2,3-b]pyran-8-yl [(2S,3R)-4-[{[2-(cyclopropylamino)-1,3-benzothiazol-6-yl]sulfonyl}(2-methylpropyl)amino]-1-(3,5-difluorophenyl)-3-hydroxybutan-2-yl]carbamate, Integrase, SULFATE ION | | Authors: | Aoki, M, Aoki-Ogata, H, Bulut, H, Hayashi, H, Davis, D, Hasegawa, K, Yarchoan, R, Ghosh, A.K, Pau, A.K, Mitsuya, H. | | Deposit date: | 2022-03-21 | | Release date: | 2023-03-22 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | GRL-142 binds to and impairs HIV-1 integrase nuclear localization signal and potently suppresses highly INSTI-resistant HIV-1 variants.
Sci Adv, 9, 2023
|
|
7RBX
 
 | | Crystal structure of isocitrate lyase and phosphorylmutase:isocitrate lyase from Brucella melitensis biovar Abortus 2308 bound to itaconic acid | | Descriptor: | 1,2-ETHANEDIOL, 2-methylidenebutanedioic acid, Isocitrase, ... | | Authors: | Seattle Structural Genomics Center for Infectious Disease, Edwards, T.E, Abendroth, J, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2021-07-06 | | Release date: | 2021-10-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Aconitate decarboxylase 1 participates in the control of pulmonary Brucella infection in mice.
Plos Pathog., 17, 2021
|
|
6E4T
 
 | | Structure of AMPK bound to activator | | Descriptor: | 1-O-{6-chloro-5-[4-(1-hydroxycyclobutyl)phenyl]-1H-indole-3-carbonyl}-beta-D-glucopyranuronic acid, 5'-AMP-activated protein kinase catalytic subunit alpha-1, 5'-AMP-activated protein kinase subunit beta-1, ... | | Authors: | Calabrese, M.F, Kurumbail, R.G. | | Deposit date: | 2018-07-18 | | Release date: | 2018-08-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Acyl Glucuronide Metabolites of 6-Chloro-5-[4-(1-hydroxycyclobutyl)phenyl]-1 H-indole-3-carboxylic Acid (PF-06409577) and Related Indole-3-carboxylic Acid Derivatives are Direct Activators of Adenosine Monophosphate-Activated Protein Kinase (AMPK).
J. Med. Chem., 61, 2018
|
|
6E4U
 
 | | Structure of AMPK bound to activator | | Descriptor: | 1-O-{6-chloro-5-[6-(dimethylamino)-2-methoxypyridin-3-yl]-1H-indole-3-carbonyl}-beta-D-glucopyranuronic acid, 5'-AMP-activated protein kinase catalytic subunit alpha-1, 5'-AMP-activated protein kinase subunit beta-1, ... | | Authors: | Calabrese, M.F, Kurumbail, R.G. | | Deposit date: | 2018-07-18 | | Release date: | 2018-08-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.27 Å) | | Cite: | Acyl Glucuronide Metabolites of 6-Chloro-5-[4-(1-hydroxycyclobutyl)phenyl]-1 H-indole-3-carboxylic Acid (PF-06409577) and Related Indole-3-carboxylic Acid Derivatives are Direct Activators of Adenosine Monophosphate-Activated Protein Kinase (AMPK).
J. Med. Chem., 61, 2018
|
|
3NL9
 
 | |
6IMU
 
 | | The apo-structure of endo-beta-1,2-glucanase from Talaromyces funiculosus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CITRIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Tanaka, N, Nakajima, M, Narukawa-Nara, M, Matsunaga, H, Kamisuki, S, Aramasa, H, Takahashi, Y, Sugimoto, N, Abe, K, Miyanaga, A, Yamashita, T, Sugawara, F, Kamakura, T, Komba, S, Nakai, H, Taguchi, H. | | Deposit date: | 2018-10-23 | | Release date: | 2019-04-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification, characterization, and structural analyses of a fungal endo-beta-1,2-glucanase reveal a new glycoside hydrolase family.
J.Biol.Chem., 294, 2019
|
|
4OCO
 
 | | N-acetylhexosamine 1-phosphate kinase in complex with GlcNAc-1-phosphate | | Descriptor: | 2-acetamido-2-deoxy-1-O-phosphono-alpha-D-glucopyranose, N-acetylhexosamine 1-phosphate kinase | | Authors: | Li, T.L, Wang, K.C, Lyu, S.Y, Liu, Y.C, Chang, C.Y, Wu, C.J. | | Deposit date: | 2014-01-09 | | Release date: | 2014-05-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Insights into the binding specificity and catalytic mechanism of N-acetylhexosamine 1-phosphate kinases through multiple reaction complexes.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3O9B
 
 | | Crystal Structure of wild-type HIV-1 Protease in Complex with kd25 | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl [(1S,2R)-1-benzyl-2-hydroxy-3-({[4-(hydroxymethyl)phenyl]sulfonyl}[(2S)-2-methylbutyl]amino)propyl]carbamate, ACETATE ION, GLYCEROL, ... | | Authors: | Schiffer, C.A, Nalam, M.N.L. | | Deposit date: | 2010-08-04 | | Release date: | 2011-08-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Substrate envelope-designed potent HIV-1 protease inhibitors to avoid drug resistance.
Chem.Biol., 20, 2013
|
|
4NJS
 
 | | Crystal structure of multidrug-resistant clinical isolate A02 HIV-1 protease in complex with non-peptidic inhibitor, GRL008 | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl [(2S,3R)-4-{[(4-carbamoylphenyl)sulfonyl](2-methylpropyl)amino}-3-hydroxy-1-phenylbutan-2-yl]carbamate, Protease | | Authors: | Yedidi, R.S, Garimella, H, Kaufman, J.D, Das, D, Wingfield, P.T, Ghosh, A.K, Mitsuya, H. | | Deposit date: | 2013-11-11 | | Release date: | 2014-04-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A Conserved Hydrogen-Bonding Network of P2 bis-Tetrahydrofuran-Containing HIV-1 Protease Inhibitors (PIs) with a Protease Active-Site Amino Acid Backbone Aids in Their Activity against PI-Resistant HIV.
Antimicrob.Agents Chemother., 58, 2014
|
|
7VIG
 
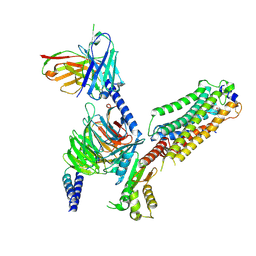 | | Cryo-EM structure of Gi coupled Sphingosine 1-phosphate receptor bound with CBP-307 | | Descriptor: | 1-[[2-fluoranyl-4-[5-[4-(2-methylpropyl)phenyl]-1,2,4-oxadiazol-3-yl]phenyl]methyl]azetidine-3-carboxylic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Yu, L.Y, Gan, B, Xiao, Q.J, Ren, R.B. | | Deposit date: | 2021-09-26 | | Release date: | 2022-09-28 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Structural insights into sphingosine-1-phosphate receptor activation.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
6H2Z
 
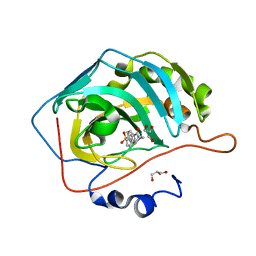 | | The crystal structure of human carbonic anhydrase II in complex with 4-(4-phenylpiperidine-1-carbonyl)benzenesulfonamide. | | Descriptor: | 4-(4-phenylpiperidin-1-yl)carbonylbenzenesulfonamide, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Buemi, M.R, Di Fiore, A, De Luca, L, Ferro, S, Mancuso, F, Monti, S.M, Buonanno, M, Angeli, A, Russo, E, De Sarro, G, Supuran, C.T, De Simone, G, Gitto, R. | | Deposit date: | 2018-07-17 | | Release date: | 2018-12-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Exploring structural properties of potent human carbonic anhydrase inhibitors bearing a 4-(cycloalkylamino-1-carbonyl)benzenesulfonamide moiety.
Eur J Med Chem, 163, 2018
|
|
