3U2B
 
 | | Structure of the Sox4 HMG domain bound to DNA | | Descriptor: | DNA (5'-D(*CP*CP*AP*GP*GP*AP*CP*AP*AP*TP*AP*GP*AP*GP*AP*C)-3'), DNA (5'-D(*GP*TP*CP*TP*CP*TP*AP*TP*TP*GP*TP*CP*CP*TP*GP*G)-3'), Transcription factor SOX-4 | | Authors: | Jauch, R, Ng, C.K.L, Kolatkar, P.R. | | Deposit date: | 2011-10-03 | | Release date: | 2011-12-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.402 Å) | | Cite: | The crystal structure of the Sox4 HMG domain-DNA complex suggests a mechanism for positional interdependence in DNA recognition
Biochem.J., 443, 2012
|
|
6KZX
 
 | | Crystal structure of E.coli DNA gyrase B in complex with 2-oxo-1,2-dihydroquinoline derivative | | Descriptor: | 3-[[8-(methylamino)-2-oxidanylidene-1~{H}-quinolin-3-yl]carbonylamino]benzoic acid, DNA gyrase subunit B | | Authors: | Mima, M, Takeuchi, T, Ushiyama, F. | | Deposit date: | 2019-09-25 | | Release date: | 2020-05-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Lead Identification of 8-(Methylamino)-2-oxo-1,2-dihydroquinoline Derivatives as DNA Gyrase Inhibitors: Hit-to-Lead Generation Involving Thermodynamic Evaluation.
Acs Omega, 5, 2020
|
|
6L01
 
 | | Crystal structure of E.coli DNA gyrase B in complex with 2-oxo-1,2-dihydroquinoline derivative | | Descriptor: | 2-[3-[[8-(methylamino)-2-oxidanylidene-1~{H}-quinolin-3-yl]carbonylamino]phenyl]ethanoic acid, DNA gyrase subunit B | | Authors: | Mima, M, Takeuchi, T, Ushiyama, F. | | Deposit date: | 2019-09-25 | | Release date: | 2020-05-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Lead Identification of 8-(Methylamino)-2-oxo-1,2-dihydroquinoline Derivatives as DNA Gyrase Inhibitors: Hit-to-Lead Generation Involving Thermodynamic Evaluation.
Acs Omega, 5, 2020
|
|
6KZZ
 
 | | Crystal structure of E.coli DNA gyrase B in complex with 2-oxo-1,2-dihydroquinoline derivative | | Descriptor: | 4-[[8-(methylamino)-2-oxidanylidene-1~{H}-quinolin-3-yl]carbonylamino]benzoic acid, DNA gyrase subunit B | | Authors: | Mima, M, Takeuchi, T, Ushiyama, F. | | Deposit date: | 2019-09-25 | | Release date: | 2020-05-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Lead Identification of 8-(Methylamino)-2-oxo-1,2-dihydroquinoline Derivatives as DNA Gyrase Inhibitors: Hit-to-Lead Generation Involving Thermodynamic Evaluation.
Acs Omega, 5, 2020
|
|
3DW9
 
 | | SgrAI with cognate DNA and manganese bound | | Descriptor: | DNA (5'-D(*DGP*DAP*DGP*DTP*DCP*DCP*DAP*DCP*DCP*DGP*DGP*DTP*DGP*DGP*DAP*DCP*DTP*DC)-3'), MANGANESE (II) ION, SgraIR restriction enzyme | | Authors: | Dunten, P.W, Horton, N.C, Little, E.J. | | Deposit date: | 2008-07-21 | | Release date: | 2008-08-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structure of SgrAI bound to DNA; recognition of an 8 base pair target.
Nucleic Acids Res., 36, 2008
|
|
142D
 
 | |
6IR8
 
 | | Rice WRKY/DNA complex | | Descriptor: | DNA (5'-D(P*GP*AP*TP*AP*TP*TP*TP*GP*AP*CP*CP*GP*GP*A)-3'), DNA (5'-D(P*TP*CP*CP*GP*GP*TP*CP*AP*AP*AP*TP*AP*TP*C)-3'), OsWRKY45, ... | | Authors: | Liu, J, Cheng, X, Wang, D. | | Deposit date: | 2018-11-12 | | Release date: | 2019-02-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis of dimerization and dual W-box DNA recognition by rice WRKY domain.
Nucleic Acids Res., 47, 2019
|
|
1ANA
 
 | |
6DSX
 
 | | Bst DNA polymerase I post-chemistry (n+1 with dATP soak) structure | | Descriptor: | DNA (5'-D(*GP*CP*GP*AP*TP*CP*AP*CP*GP*TP*A)-3'), DNA (5'-D(*GP*TP*AP*CP*GP*TP*GP*AP*TP*CP*GP*CP*A)-3'), DNA polymerase I, ... | | Authors: | Chim, N, Jackson, L.N, Chaput, J.C. | | Deposit date: | 2018-06-14 | | Release date: | 2018-10-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Crystal structures of DNA polymerase I capture novel intermediates in the DNA synthesis pathway.
Elife, 7, 2018
|
|
6R92
 
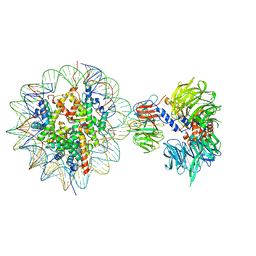 | | Cryo-EM structure of NCP-THF2(+1)-UV-DDB class B | | Descriptor: | DNA damage-binding protein 1,DNA damage-binding protein 1, DNA damage-binding protein 2, Histone H2A type 1-B/E, ... | | Authors: | Matsumoto, S, Cavadini, S, Bunker, R.D, Thoma, N.H. | | Deposit date: | 2019-04-02 | | Release date: | 2019-06-12 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | DNA damage detection in nucleosomes involves DNA register shifting.
Nature, 571, 2019
|
|
1LAT
 
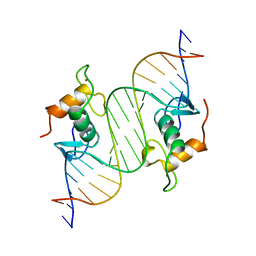 | | GLUCOCORTICOID RECEPTOR MUTANT/DNA COMPLEX | | Descriptor: | DNA (5'-D(*TP*TP*CP*CP*AP*GP*AP*AP*CP*AP*TP*GP*TP*TP*CP*TP*G P*GP*A)-3'), GLUCOCORTICOID RECEPTOR, ZINC ION | | Authors: | Gewirth, D.T, Sigler, P.B. | | Deposit date: | 1995-12-18 | | Release date: | 1996-04-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The basis for half-site specificity explored through a non-cognate steroid receptor-DNA complex.
Nat.Struct.Biol., 2, 1995
|
|
2BOP
 
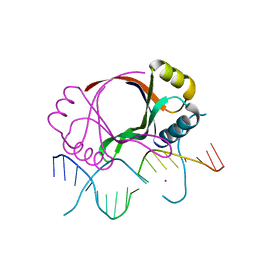 | | CRYSTAL STRUCTURE AT 1.7 ANGSTROMS OF THE BOVINE PAPILLOMAVIRUS-1 E2 DNA-BINDING DOMAIN BOUND TO ITS DNA TARGET | | Descriptor: | DNA (5'-D(*CP*CP*GP*AP*CP*CP*GP*AP*CP*GP*TP*CP*GP*GP*TP*CP*G )-3'), PROTEIN (E2), YTTERBIUM (III) ION | | Authors: | Hegde, R.S, Grossman, S.R, Laimins, L.A, Sigler, P.B. | | Deposit date: | 1994-01-13 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure at 1.7 A of the bovine papillomavirus-1 E2 DNA-binding domain bound to its DNA target.
Nature, 359, 1992
|
|
7ZTH
 
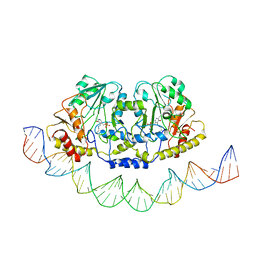 | | Cryo-EM structure of holo-PdxR from Bacillus clausii bound to its target DNA in the open conformation | | Descriptor: | DNA (48-MER), PLP-dependent aminotransferase family protein | | Authors: | Freda, I, Montemiglio, L.C, Tramonti, A, Contestabile, R, Vallone, B, Exertier, C, Savino, C, Chaves Sanjuan, A, Bolognesi, M. | | Deposit date: | 2022-05-10 | | Release date: | 2023-07-05 | | Last modified: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural insights into the DNA recognition mechanism by the bacterial transcription factor PdxR.
Nucleic Acids Res., 51, 2023
|
|
7ZPA
 
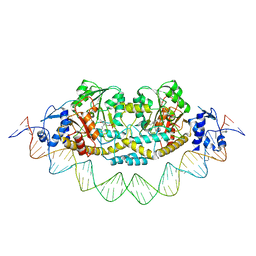 | | Cryo-EM structure of holo-PdxR from Bacillus clausii bound to its target DNA in the closed conformation, C1 symmetry | | Descriptor: | DNA (48-MER), PLP-dependent aminotransferase family protein | | Authors: | Freda, I, Montemiglio, L.C, Tramonti, A, Contestabile, R, Vallone, B, Exertier, C, Savino, C, Chaves Sanjuan, A, Bolognesi, M. | | Deposit date: | 2022-04-27 | | Release date: | 2023-07-05 | | Last modified: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural insights into the DNA recognition mechanism by the bacterial transcription factor PdxR.
Nucleic Acids Res., 51, 2023
|
|
7ZLA
 
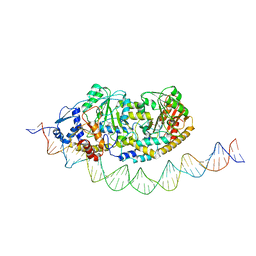 | | Cryo-EM structure of holo-PdxR from Bacillus clausii bound to its target DNA in the half-closed conformation | | Descriptor: | DNA (48-MER), PLP-dependent aminotransferase family protein | | Authors: | Freda, I, Montemiglio, L.C, Tramonti, A, Contestabile, R, Vallone, B, Savino, C, Exertier, C, Bolognesi, M, Chaves Sanjuan, A. | | Deposit date: | 2022-04-14 | | Release date: | 2023-07-05 | | Last modified: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (3.99 Å) | | Cite: | Structural insights into the DNA recognition mechanism by the bacterial transcription factor PdxR.
Nucleic Acids Res., 51, 2023
|
|
4OWX
 
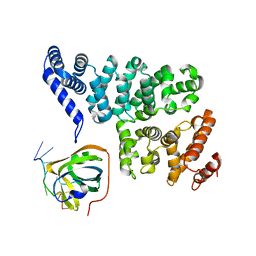 | | Structural basis of SOSS1 in complex with a 12nt ssDNA | | Descriptor: | DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), Integrator complex subunit 3, SOSS complex subunit B1 | | Authors: | Ren, W, Sun, Q, Tang, X, Song, H. | | Deposit date: | 2014-02-04 | | Release date: | 2014-04-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis of SOSS1 Complex Assembly and Recognition of ssDNA.
Cell Rep, 6, 2014
|
|
8VX6
 
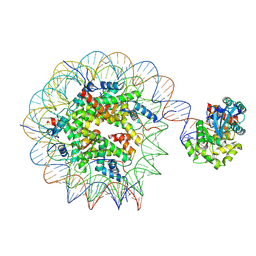 | | Human OGG1 bound at the nucleosomal DNA entry site | | Descriptor: | DNA (167-MER), Histone H2A, Histone H2B 1.1, ... | | Authors: | You, Q, Li, H. | | Deposit date: | 2024-02-03 | | Release date: | 2024-10-09 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Human 8-oxoguanine glycosylase OGG1 binds nucleosome at the dsDNA ends and the super-helical locations.
Commun Biol, 7, 2024
|
|
8VX4
 
 | | Human OGG1 bound to a 35-bp DNA with an 8-oxoG in the middle | | Descriptor: | DNA (35-MER), N-glycosylase/DNA lyase | | Authors: | You, Q, Li, H. | | Deposit date: | 2024-02-03 | | Release date: | 2024-10-09 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Human 8-oxoguanine glycosylase OGG1 binds nucleosome at the dsDNA ends and the super-helical locations.
Commun Biol, 7, 2024
|
|
5T01
 
 | | Human c-Jun DNA binding domain homodimer in complex with methylated DNA | | Descriptor: | DNA (5'-D(*AP*AP*TP*GP*GP*AP*(5CM)P*GP*AP*GP*TP*CP*AP*TP*AP*GP*GP*AP*G)-3'), DNA (5'-D(P*CP*TP*CP*CP*TP*AP*TP*GP*AP*CP*TP*CP*GP*TP*CP*CP*AP*T)-3'), Transcription factor AP-1 | | Authors: | Hong, S, Horton, J.R, Cheng, X. | | Deposit date: | 2016-08-15 | | Release date: | 2017-03-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Methyl-dependent and spatial-specific DNA recognition by the orthologous transcription factors human AP-1 and Epstein-Barr virus Zta.
Nucleic Acids Res., 45, 2017
|
|
5DDM
 
 | |
1BDZ
 
 | |
6DSV
 
 | | Bst DNA polymerase I post-chemistry (n+2) structure | | Descriptor: | DNA (5'-D(*GP*CP*GP*AP*TP*CP*AP*CP*GP*TP*A)-3'), DNA (5'-D(P*TP*AP*CP*GP*TP*GP*AP*TP*CP*GP*CP*A)-3'), DNA polymerase I, ... | | Authors: | Chim, N, Jackson, L.N, Chaput, J.C. | | Deposit date: | 2018-06-14 | | Release date: | 2018-10-31 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Crystal structures of DNA polymerase I capture novel intermediates in the DNA synthesis pathway.
Elife, 7, 2018
|
|
6ON0
 
 | | STRUCTURE OF N15 CRO COMPLEXED WITH CONSENSUS OPERATOR DNA | | Descriptor: | DNA (5'-D(*TP*TP*TP*AP*TP*AP*GP*CP*TP*AP*GP*CP*TP*AP*TP*AP*A)-3'), Gp39 | | Authors: | Hall, B.M, Roberts, S.A, Cordes, M.H.J. | | Deposit date: | 2019-04-19 | | Release date: | 2019-05-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Extreme divergence between one-to-one orthologs: the structure of N15 Cro bound to operator DNA and its relationship to the lambda Cro complex.
Nucleic Acids Res., 47, 2019
|
|
6DSY
 
 | | Bst DNA polymerase I post-chemistry (n+1) structure | | Descriptor: | DNA (5'-D(*GP*CP*GP*AP*TP*CP*AP*CP*GP*T)-3'), DNA (5'-D(P*GP*TP*AP*CP*GP*TP*GP*AP*TP*CP*GP*CP*A)-3'), DNA polymerase I, ... | | Authors: | Chim, N, Jackson, L.N, Chaput, J.C. | | Deposit date: | 2018-06-14 | | Release date: | 2018-10-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Crystal structures of DNA polymerase I capture novel intermediates in the DNA synthesis pathway.
Elife, 7, 2018
|
|
1C32
 
 | | SOLUTION STRUCTURE OF A QUADRUPLEX FORMING DNA AND ITS INTERMIDIATE | | Descriptor: | DNA (5'-D(*GP*GP*TP*TP*GP*GP*TP*GP*TP*GP*GP*TP*TP*GP*G)-3') | | Authors: | Bolton, P.H, Marathias, V.M, Wang, K. | | Deposit date: | 1999-07-24 | | Release date: | 1999-08-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structures of the potassium-saturated, 2:1, and intermediate, 1:1, forms of a quadruplex DNA.
Nucleic Acids Res., 28, 2000
|
|
