7Q61
 
 | | Structure of TEV conjugated A2ML1 (A2ML1-TC) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-2-macroglobulin-like protein 1 | | Authors: | Nielsen, N.S, Zarantonello, A, Andersen, G.R. | | Deposit date: | 2021-11-05 | | Release date: | 2022-04-20 | | Last modified: | 2022-11-02 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | Cryo-EM structures of human A2ML1 elucidate the protease-inhibitory mechanism of the A2M family.
Nat Commun, 13, 2022
|
|
6V7F
 
 | | Human Arginase1 Complexed with Bicyclic Inhibitor Compound 13 | | Descriptor: | Arginase-1, MANGANESE (II) ION, {3-[(5R,7S,8S)-8-azaniumyl-8-carboxy-2-azaspiro[4.4]nonan-2-ium-7-yl]propyl}(trihydroxy)borate(1-) | | Authors: | Palte, R.L, Lesburg, C.A. | | Deposit date: | 2019-12-08 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Discovery and Optimization of Rationally Designed Bicyclic Inhibitors of Human Arginase to Enhance Cancer Immunotherapy.
Acs Med.Chem.Lett., 11, 2020
|
|
7PWO
 
 | | Cryo-EM structure of Giardia lamblia ribosome at 2.75 A resolution | | Descriptor: | 40S ribosomal protein S21, 40S ribosomal protein S26, 40S ribosomal protein S30, ... | | Authors: | Hiregange, D.G, Rivalta, A, Bose, T, Breiner-Goldstein, E, Samiya, S, Cimicata, G, Kulakova, L, Zimmerman, E, Bashan, A, Herzberg, O, Yonath, A. | | Deposit date: | 2021-10-07 | | Release date: | 2022-04-20 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.75 Å) | | Cite: | Cryo-EM structure of the ancient eukaryotic ribosome from the human parasite Giardia lamblia.
Nucleic Acids Res., 50, 2022
|
|
8D13
 
 | | Helical ADP-F-actin | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Reynolds, M.J, Alushin, G.M. | | Deposit date: | 2022-05-26 | | Release date: | 2022-09-07 | | Last modified: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (2.43 Å) | | Cite: | Bending forces and nucleotide state jointly regulate F-actin structure.
Nature, 611, 2022
|
|
4PY3
 
 | |
7ND2
 
 | | Cryo-EM structure of the human FERRY complex | | Descriptor: | Glutamine amidotransferase-like class 1 domain-containing protein 1, Protein phosphatase 1 regulatory subunit 21, Quinone oxidoreductase-like protein 1 | | Authors: | Quentin, D, Klink, B.U, Raunser, S. | | Deposit date: | 2021-01-29 | | Release date: | 2022-03-02 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural basis of mRNA binding by the human FERRY Rab5 effector complex.
Mol.Cell, 83, 2023
|
|
6AQ1
 
 | | The crystal structure of human FABP3 | | Descriptor: | Fatty acid-binding protein, heart, PALMITIC ACID, ... | | Authors: | Hsu, H.C, Li, H. | | Deposit date: | 2017-08-18 | | Release date: | 2018-06-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.40000093 Å) | | Cite: | SAR studies on truxillic acid mono esters as a new class of antinociceptive agents targeting fatty acid binding proteins.
Eur J Med Chem, 154, 2018
|
|
5IRU
 
 | | Crystal structure of avidin in complex with 1-biotinylpyrene | | Descriptor: | 1-biotinylpyrene, 2-acetamido-2-deoxy-beta-D-glucopyranose, Avidin | | Authors: | Strzelczyk, P, Bujacz, G. | | Deposit date: | 2016-03-14 | | Release date: | 2016-10-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Characterization of the Avidin Interactions with Fluorescent Pyrene-Conjugates: 1-Biotinylpyrene and 1-Desthiobiotinylpyrene.
Molecules, 21, 2016
|
|
5IS1
 
 | |
8D17
 
 | | Straight ADP-F-actin 1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Reynolds, M.J, Alushin, G.M. | | Deposit date: | 2022-05-26 | | Release date: | 2022-09-07 | | Last modified: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (3.69 Å) | | Cite: | Bending forces and nucleotide state jointly regulate F-actin structure.
Nature, 611, 2022
|
|
8D16
 
 | | Bent ADP-Pi-F-actin | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Reynolds, M.J, Alushin, G.M. | | Deposit date: | 2022-05-26 | | Release date: | 2022-09-07 | | Last modified: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (3.71 Å) | | Cite: | Bending forces and nucleotide state jointly regulate F-actin structure.
Nature, 611, 2022
|
|
8CZI
 
 | |
7Q52
 
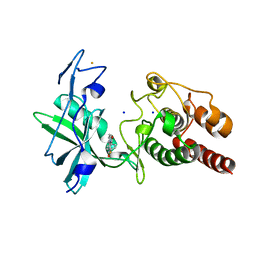 | | Crystal structure of S/T protein kinase PknG from Mycobacterium tuberculosis in complex with inhibitor L2W | | Descriptor: | 2-azanyl-3-(4-fluorophenyl)carbonyl-indolizine-1-carboxamide, FE (III) ION, SODIUM ION, ... | | Authors: | Defelipe, L.A, Burastero, O, Bento, I, Garcia-Alai, M.M. | | Deposit date: | 2021-11-02 | | Release date: | 2022-06-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Cosolvent Sites-Based Discovery of Mycobacterium Tuberculosis Protein Kinase G Inhibitors.
J.Med.Chem., 65, 2022
|
|
3IJO
 
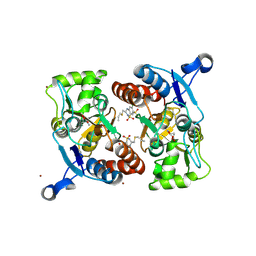 | | Crystal structure of the AMPA subunit GluR2 bound to the allosteric modulator, althiazide | | Descriptor: | (3S)-6-chloro-3-[(prop-2-en-1-ylsulfanyl)methyl]-3,4-dihydro-2H-1,2,4-benzothiadiazine-7-sulfonamide 1,1-dioxide, GLUTAMIC ACID, Glutamate receptor 2, ... | | Authors: | Ptak, C.P, Ahmed, A.H, Oswald, R.E. | | Deposit date: | 2009-08-04 | | Release date: | 2009-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.003 Å) | | Cite: | Probing the allosteric modulator binding site of GluR2 with thiazide derivatives
Biochemistry, 48, 2009
|
|
7NPA
 
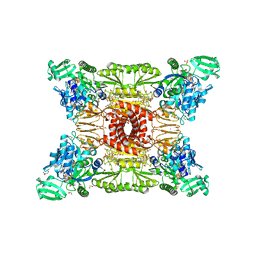 | |
6B7D
 
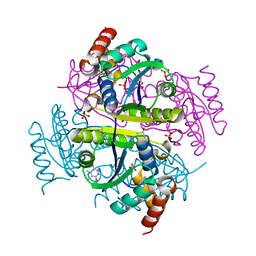 | | Crystal structure of E.coli Phosphopantetheine Adenylyltransferase (PPAT/CoaD) in complex with 3-(4-chlorophenyl)-6-methoxy-4,5-dimethylpyridazine | | Descriptor: | 3-(4-chlorophenyl)-6-methoxy-4,5-dimethylpyridazine, DIMETHYL SULFOXIDE, POTASSIUM ION, ... | | Authors: | Proudfoot, A.W, Bussiere, D, Lingel, A. | | Deposit date: | 2017-10-03 | | Release date: | 2017-12-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High-Confidence Protein-Ligand Complex Modeling by NMR-Guided Docking Enables Early Hit Optimization.
J. Am. Chem. Soc., 139, 2017
|
|
7PG5
 
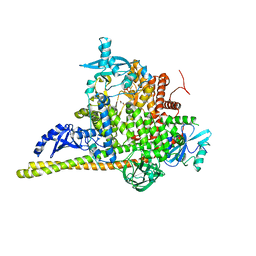 | | Crystal Structure of PI3Kalpha | | Descriptor: | GLYCEROL, PHOSPHATE ION, Phosphatidylinositol 3-kinase regulatory subunit alpha, ... | | Authors: | Gong, G, Pinotsis, N, Williams, R.L, Vanhaesebroeck, B. | | Deposit date: | 2021-08-13 | | Release date: | 2022-08-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.20029068 Å) | | Cite: | A small-molecule PI3K alpha activator for cardioprotection and neuroregeneration.
Nature, 618, 2023
|
|
7NP8
 
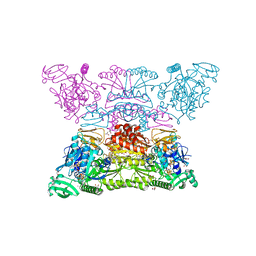 | |
6B7C
 
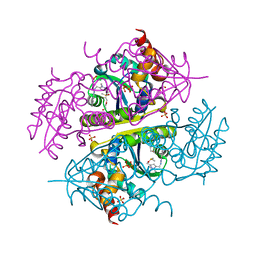 | | Crystal structure of E.coli Phosphopantetheine Adenylyltransferase (PPAT/CoaD) in complex with N-((1,3-dimethyl-1H-pyrazol-5-yl)methyl)-5-methyl-1H-imidazo[4,5-b]pyridin-2-amine | | Descriptor: | DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, N-[(1,3-dimethyl-1H-pyrazol-5-yl)methyl]-5-methyl-3H-imidazo[4,5-b]pyridin-2-amine, ... | | Authors: | Proudfoot, A.W, Bussiere, D, Lingel, A. | | Deposit date: | 2017-10-03 | | Release date: | 2017-12-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.564 Å) | | Cite: | High-Confidence Protein-Ligand Complex Modeling by NMR-Guided Docking Enables Early Hit Optimization.
J. Am. Chem. Soc., 139, 2017
|
|
7PG6
 
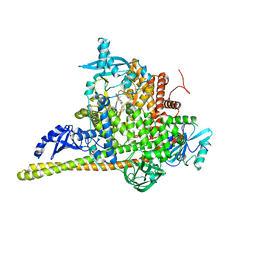 | | Crystal Structure of PI3Kalpha in complex with the inhibitor NVP-BYL719 | | Descriptor: | (2S)-N~1~-{4-methyl-5-[2-(1,1,1-trifluoro-2-methylpropan-2-yl)pyridin-4-yl]-1,3-thiazol-2-yl}pyrrolidine-1,2-dicarboxamide, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform, ... | | Authors: | Gong, G, Pinotsis, N, Williams, R.L, Vanhaesebroeck, B. | | Deposit date: | 2021-08-13 | | Release date: | 2022-08-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.49943733 Å) | | Cite: | A small-molecule PI3K alpha activator for cardioprotection and neuroregeneration.
Nature, 618, 2023
|
|
1F44
 
 | | CRYSTAL STRUCTURE OF TRIMERIC CRE RECOMBINASE-LOX COMPLEX | | Descriptor: | CRE RECOMBINASE, DNA (5'- D(*TP*AP*TP*AP*AP*CP*TP*TP*CP*GP*TP*AP*TP*AP*GP*C)-3'), DNA (5'-D(*AP*TP*AP*TP*GP*CP*TP*AP*TP*AP*CP*GP*AP*AP*GP*TP*TP*AP*T)-3') | | Authors: | Baldwin, E.P, Woods, K.C. | | Deposit date: | 2000-06-07 | | Release date: | 2001-10-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Quasi-equivalence in site-specific recombinase structure and function: crystal structure and activity of trimeric Cre recombinase bound to a three-way Lox DNA junction.
J.Mol.Biol., 313, 2001
|
|
6B3S
 
 | |
5XYW
 
 | |
6B7A
 
 | | Crystal structure of E.coli Phosphopantetheine Adenylyltransferase (PPAT/CoaD) in complex with 2-methyl-1H-benzo[d]imidazol-4-ol | | Descriptor: | 2-methyl-1H-benzimidazol-7-ol, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Proudfoot, A.W, Bussiere, D, Lingel, A. | | Deposit date: | 2017-10-03 | | Release date: | 2017-12-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.991 Å) | | Cite: | High-Confidence Protein-Ligand Complex Modeling by NMR-Guided Docking Enables Early Hit Optimization.
J. Am. Chem. Soc., 139, 2017
|
|
6VHA
 
 | | Singlet Tau Fibril from Corticobasal Degeneration Human Brain Tissue | | Descriptor: | Microtubule-associated protein tau | | Authors: | Arakhamia, T, Lee, C.E, Carlomagno, Y, Duong, D.M, Kundinger, S.R, Wang, K, Williams, D, DeTure, M, Dickson, D.W, Cook, C.N, Seyfried, N.T, Petrucelli, L, Fitzpatrick, A.W.P. | | Deposit date: | 2020-01-09 | | Release date: | 2020-03-04 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Posttranslational Modifications Mediate the Structural Diversity of Tauopathy Strains.
Cell, 180, 2020
|
|
