6LCN
 
 | | Crystal structure of Serine Acetyltransferase from Planctomyces limnophilus at 2.15A | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Kumar, N, Singh, R.P, Singh, A.K, Kumaran, S. | | Deposit date: | 2019-11-19 | | Release date: | 2020-12-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Understanding Mechanics of competitive-allostery Using Engineered Cysteine Synthase Assembly
To Be Published
|
|
2HYY
 
 | | Human Abl kinase domain in complex with imatinib (STI571, Glivec) | | Descriptor: | 4-(4-METHYL-PIPERAZIN-1-YLMETHYL)-N-[4-METHYL-3-(4-PYRIDIN-3-YL-PYRIMIDIN-2-YLAMINO)-PHENYL]-BENZAMIDE, Proto-oncogene tyrosine-protein kinase ABL1 | | Authors: | Cowan-Jacob, S.W, Fendrich, G, Liebetanz, J, Fabbro, D, Manley, P. | | Deposit date: | 2006-08-08 | | Release date: | 2007-01-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural biology contributions to the discovery of drugs to treat chronic myelogenous leukaemia.
ACTA CRYSTALLOGR.,SECT.D, 63, 2007
|
|
6LD6
 
 | |
6LE1
 
 | | Structure of RRM2 domain of DND1 protein | | Descriptor: | Dead end protein homolog 1, GLYCEROL, SULFATE ION | | Authors: | Kumari, P, Bhavesh, N.S. | | Deposit date: | 2019-11-23 | | Release date: | 2020-12-02 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Human DND1-RRM2 forms a non-canonical domain swapped dimer.
Protein Sci., 30, 2021
|
|
2HZK
 
 | | Crystal structures of a sodium-alpha-keto acid binding subunit from a TRAP transporter in its open form | | Descriptor: | GLYCEROL, TRAP-T family sorbitol/mannitol transporter, periplasmic binding protein, ... | | Authors: | Gonin, S, Arnoux, P, Pierru, B, Alonso, B, Sabaty, M, Pignol, D. | | Deposit date: | 2006-08-09 | | Release date: | 2007-04-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of an Extracytoplasmic Solute Receptor from a TRAP transporter in its open and closed forms reveal a helix-swapped dimer requiring a cation for alpha-keto acid binding.
Bmc Struct.Biol., 7, 2007
|
|
6LEH
 
 | | Crystal structure of Autotaxin in complex with an inhibitor | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Nishimasu, H, Osamu, N. | | Deposit date: | 2019-11-25 | | Release date: | 2020-03-18 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification of PotentIn VivoAutotaxin Inhibitors that Bind to Both Hydrophobic Pockets and Channels in the Catalytic Domain.
J.Med.Chem., 63, 2020
|
|
6L96
 
 | | Structure of PPARalpha-LBD/pemafibrate/SRC1 peptide | | Descriptor: | (2~{R})-2-[3-[[1,3-benzoxazol-2-yl-[3-(4-methoxyphenoxy)propyl]amino]methyl]phenoxy]butanoic acid, Peroxisome proliferator-activated receptor alpha, SRC1 coactivator peptide | | Authors: | Kawasaki, M, Kambe, A, Yamamoto, Y, Arulmozhira, S, Ito, S, Nakagawa, Y, Tokiwa, H, Nakano, S, Shimano, H. | | Deposit date: | 2019-11-08 | | Release date: | 2020-01-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Elucidation of Molecular Mechanism of a Selective PPAR alpha Modulator, Pemafibrate, through Combinational Approaches of X-ray Crystallography, Thermodynamic Analysis, and First-Principle Calculations.
Int J Mol Sci, 21, 2020
|
|
6L9L
 
 | | 1D4 TCR recognition of H2-Ld a1a2 A5 Peptide Complexes | | Descriptor: | H2-Ld a1a2, SER-PRO-SER-TYR-ALA-TYR-HIS-GLN-PHE, T Cell Receptor | | Authors: | Wei, P.C, Yin, L. | | Deposit date: | 2019-11-10 | | Release date: | 2020-11-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.399 Å) | | Cite: | Structures suggest an approach for converting weak self-peptide tumor antigens into superagonists for CD8 T cells in cancer.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6LEV
 
 | | Quadruple mutant (N51I+C59R+S108N+I164L) plasmodium falciparum dihydrofolate reductase-thymidylate synthase (PfDHFR-TS) complexed with compound 46 and NADPH | | Descriptor: | 2-[[4,6-bis(azanyl)-2,2-dimethyl-1,3,5-triazin-1-yl]oxy]-N-(4-chlorophenyl)ethanamide, Bifunctional dihydrofolate reductase-thymidylate synthase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Vanichtanankul, J, Vitsupakorn, D. | | Deposit date: | 2019-11-27 | | Release date: | 2020-12-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.644 Å) | | Cite: | Flexible diaminodihydrotriazine inhibitors of Plasmodium falciparum dihydrofolate reductase: Binding strengths, modes of binding and their antimalarial activities.
Eur.J.Med.Chem., 195, 2020
|
|
2I0G
 
 | | Benzopyrans are Selective Estrogen Receptor beta Agonists (SERBAs) with Novel Activity in Models of Benign Prostatic Hyperplasia | | Descriptor: | (3AS,4R,9BR)-4-(4-HYDROXYPHENYL)-1,2,3,3A,4,9B-HEXAHYDROCYCLOPENTA[C]CHROMEN-8-OL, Estrogen receptor beta | | Authors: | Wang, Y. | | Deposit date: | 2006-08-10 | | Release date: | 2006-10-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Benzopyrans are selective estrogen receptor Beta agonists with novel activity in models of benign prostatic hyperplasia.
J.Med.Chem., 49, 2006
|
|
6LFB
 
 | | E. coli Thioesterase I mutant DG | | Descriptor: | Acyl-CoA thioesterase I also functions as protease I | | Authors: | Deng, X, Chen, L, Yang, G. | | Deposit date: | 2019-12-01 | | Release date: | 2020-07-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structure-guided reshaping of the acyl binding pocket of 'TesA thioesterase enhances octanoic acid production in E. coli.
Metab. Eng., 61, 2020
|
|
6LAD
 
 | |
6LB2
 
 | | Crystal structure of rhesus macaque MHC class I molecule Mamu-B*098 complexed with mono-acyl glycerol | | Descriptor: | (2R)-2,3-dihydroxypropyl hexadecanoate, 1,2-ETHANEDIOL, Beta-2-microglobulin, ... | | Authors: | Shima, Y, Morita, D. | | Deposit date: | 2019-11-13 | | Release date: | 2020-04-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.69380951 Å) | | Cite: | Crystal structures of lysophospholipid-bound MHC class I molecules.
J.Biol.Chem., 295, 2020
|
|
6LG4
 
 | |
6LGA
 
 | | Bombyx mori GH13 sucrose hydrolase | | Descriptor: | CALCIUM ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Miyazaki, T. | | Deposit date: | 2019-12-05 | | Release date: | 2020-05-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure-function analysis of silkworm sucrose hydrolase uncovers the mechanism of substrate specificity in GH13 subfamily 17exo-alpha-glucosidases.
J.Biol.Chem., 295, 2020
|
|
6LGO
 
 | |
2I6S
 
 | | Complement component C2a | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Complement C2a fragment, ... | | Authors: | Milder, F.J, Raaijmakers, H.C.A, Vandeputte, D.A.A, Schouten, A, Huizinga, E.G, Romijn, R.A, Hemrika, W, Roos, A, Daha, M.R, Gros, P. | | Deposit date: | 2006-08-29 | | Release date: | 2006-10-17 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of complement component c2a: implications for convertase formation and substrate binding.
Structure, 14, 2006
|
|
6LHS
 
 | | High resolution structure of FANCA C-terminal domain (CTD) | | Descriptor: | Fanconi anemia complementation group A | | Authors: | Jeong, E, Lee, S, Shin, J, Kim, Y, Scharer, O, Kim, Y, Kim, H, Cho, Y. | | Deposit date: | 2019-12-10 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Structural basis of the fanconi anemia-associated mutations within the FANCA and FANCG complex.
Nucleic Acids Res., 48, 2020
|
|
6LBL
 
 | | Crystal structure of IMP-1 metallo-beta-lactamase in complex with NO9 inhibitor | | Descriptor: | 2,5-dimethyl-4-sulfamoyl-furan-3-carboxylic acid, Metallo-beta-lactamase type 2, SODIUM ION, ... | | Authors: | Wachino, J. | | Deposit date: | 2019-11-14 | | Release date: | 2020-02-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Sulfamoyl Heteroarylcarboxylic Acids as Promising Metallo-beta-Lactamase Inhibitors for Controlling Bacterial Carbapenem Resistance.
Mbio, 11, 2020
|
|
6LBQ
 
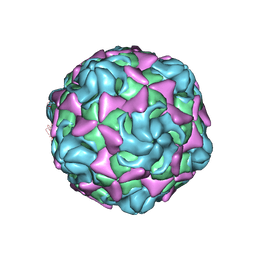 | | Cryo-EM structure of echovirus 11 empty particle at pH 5.5 | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3 | | Authors: | Liu, S, Gao, F.G. | | Deposit date: | 2019-11-14 | | Release date: | 2020-10-07 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Molecular and structural basis of Echovirus 11 infection by using the dual-receptor system of CD55 and FcRn.
Chin.Sci.Bull., 2020
|
|
6LCM
 
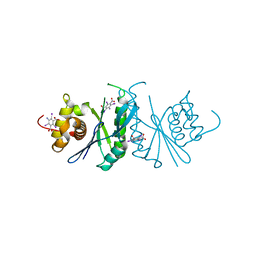 | | Crystal structure of chloroplast resolvase ZmMOC1 with the magic triangle I3C | | Descriptor: | 5-amino-2,4,6-triiodobenzene-1,3-dicarboxylic acid, ZmMoc1 | | Authors: | Yan, J.J, Hong, S.X, Guan, Z.Y, Yin, P. | | Deposit date: | 2019-11-19 | | Release date: | 2020-04-08 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into sequence-dependent Holliday junction resolution by the chloroplast resolvase MOC1.
Nat Commun, 11, 2020
|
|
6LIU
 
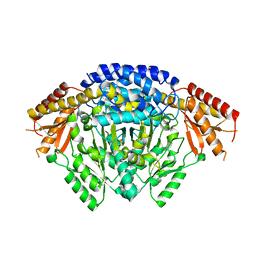 | | Crystal structure of apo Tyrosine decarboxylase | | Descriptor: | Tyrosine/DOPA decarboxylase 2 | | Authors: | Yu, J, Wang, H, Yao, M. | | Deposit date: | 2019-12-13 | | Release date: | 2020-02-12 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures clarify cofactor binding of plant tyrosine decarboxylase.
Biochem.Biophys.Res.Commun., 2019
|
|
6LKC
 
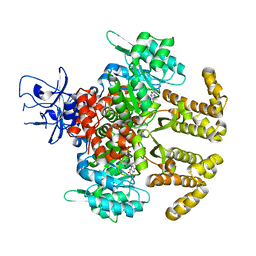 | | Crystal structure of PfaD from Shewanella piezotolerans in complex with FMN | | Descriptor: | CALCIUM ION, FLAVIN MONONUCLEOTIDE, GLYCEROL, ... | | Authors: | Zhang, M.L, Li, Q, Meng, S.S, Guo, L.J, He, L, Huang, J.Z, Li, L, Zhang, H.D. | | Deposit date: | 2019-12-19 | | Release date: | 2020-12-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | Structural Insights into the Trans -Acting Enoyl Reductase in the Biosynthesis of Long-Chain Polyunsaturated Fatty Acids in Shewanella piezotolerans .
J.Agric.Food Chem., 69, 2021
|
|
6KPL
 
 | | Crystal Structure of endo-beta-N-acetylglucosaminidase from Cordyceps militaris in apo form | | Descriptor: | Chitinase, DI(HYDROXYETHYL)ETHER, TRIETHYLENE GLYCOL | | Authors: | Seki, H, Arakawa, T, Yamada, C, Takegawa, K, Fushinobu, S. | | Deposit date: | 2019-08-15 | | Release date: | 2019-10-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis for the specific cleavage of core-fucosylatedN-glycans by endo-beta-N-acetylglucosaminidase from the fungusCordyceps militaris.
J.Biol.Chem., 294, 2019
|
|
2I4T
 
 | | Crystal structure of Purine Nucleoside Phosphorylase from Trichomonas vaginalis with Imm-A | | Descriptor: | 3,4-PYRROLIDINEDIOL,2-(4-AMINO-5H-PYRROLO[3,2-D]PYRIMIDIN-7-YL)-5-(HYDROXYMETHYL)-2S,3S,4R,5R, PHOSPHATE ION, Trichomonas vaginalis purine nucleoside phosphorylase | | Authors: | Rinaldo-Matthis, A, Schramm, V.L, Almo, S.C. | | Deposit date: | 2006-08-22 | | Release date: | 2007-06-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Inhibition and structure of Trichomonas vaginalis purine nucleoside phosphorylase with picomolar transition state analogues.
Biochemistry, 46, 2007
|
|
