6XXU
 
 | | Solution NMR structure of the native form of UbcH7 (UBE2L3) | | Descriptor: | Ubiquitin-conjugating enzyme E2 L3 | | Authors: | Marousis, K.D, Seliami, A, Birkou, M, Episkopou, V, Spyroulias, G.A. | | Deposit date: | 2020-01-28 | | Release date: | 2020-02-05 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | 1H,13C,15N backbone and side-chain resonance assignment of the native form of UbcH7 (UBE2L3) through solution NMR spectroscopy.
Biomol.Nmr Assign., 14, 2020
|
|
6ULM
 
 | | Crystal structure of human cadherin 17 EC1-2 | | Descriptor: | CALCIUM ION, Cadherin-17 | | Authors: | Gray, M.E, Sotomayor, M. | | Deposit date: | 2019-10-08 | | Release date: | 2021-03-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of the nonclassical cadherin-17 N-terminus and implications for its adhesive binding mechanism.
Acta Crystallogr.,Sect.F, 77, 2021
|
|
7PK9
 
 | | C-reactive protein decamer at pH 7.5 | | Descriptor: | C-reactive protein, CALCIUM ION | | Authors: | Noone, D.P, Sharp, T.H. | | Deposit date: | 2021-08-25 | | Release date: | 2021-12-22 | | Last modified: | 2022-01-12 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Cryo-Electron Microscopy and Biochemical Analysis Offer Insights Into the Effects of Acidic pH, Such as Occur During Acidosis, on the Complement Binding Properties of C-Reactive Protein.
Front Immunol, 12, 2021
|
|
6XZ7
 
 | | E. coli 50S ribosomal subunit in complex with dirithromycin, fMet-Phe-tRNA(Phe) and deacylated tRNA(iMet). | | Descriptor: | 23S rRNA, 50S ribosomal protein L10, 50S ribosomal protein L11, ... | | Authors: | Pichkur, E.B, Polikanov, Y.S, Myasnikov, A.G, Konevega, A.L. | | Deposit date: | 2020-02-03 | | Release date: | 2020-07-22 | | Method: | ELECTRON MICROSCOPY (2.1 Å) | | Cite: | Insights into the improved macrolide inhibitory activity from the high-resolution cryo-EM structure of dirithromycin bound to theE. coli70S ribosome.
Rna, 26, 2020
|
|
7PKB
 
 | | C-reactive protein pentamer at pH 7.5 | | Descriptor: | C-reactive protein, CALCIUM ION | | Authors: | Noone, D.P, Sharp, T.H. | | Deposit date: | 2021-08-25 | | Release date: | 2021-12-22 | | Last modified: | 2022-01-12 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-Electron Microscopy and Biochemical Analysis Offer Insights Into the Effects of Acidic pH, Such as Occur During Acidosis, on the Complement Binding Properties of C-Reactive Protein.
Front Immunol, 12, 2021
|
|
7PKF
 
 | | C-reactive protein decamer at pH 5 | | Descriptor: | C-reactive protein, CALCIUM ION | | Authors: | Noone, D.P, Sharp, T.H. | | Deposit date: | 2021-08-25 | | Release date: | 2021-12-22 | | Last modified: | 2022-01-12 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Cryo-Electron Microscopy and Biochemical Analysis Offer Insights Into the Effects of Acidic pH, Such as Occur During Acidosis, on the Complement Binding Properties of C-Reactive Protein.
Front Immunol, 12, 2021
|
|
7PKE
 
 | | C-reactive protein pentamer at pH 7.5 with phosphocholine ligand | | Descriptor: | C-reactive protein, CALCIUM ION, PHOSPHOCHOLINE | | Authors: | Noone, D.P, Sharp, T.H. | | Deposit date: | 2021-08-25 | | Release date: | 2021-12-22 | | Last modified: | 2022-01-12 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-Electron Microscopy and Biochemical Analysis Offer Insights Into the Effects of Acidic pH, Such as Occur During Acidosis, on the Complement Binding Properties of C-Reactive Protein.
Front Immunol, 12, 2021
|
|
6A6R
 
 | | Crystal structure of the modified fructosyl peptide oxidase from Aspergillus nidulans, Seleno-methionine Derivative | | Descriptor: | (4S,5S)-1,2-DITHIANE-4,5-DIOL, FLAVIN-ADENINE DINUCLEOTIDE, Fructosyl amine: oxygen oxidoreductase, ... | | Authors: | Ogawa, N, Maruyama, Y, Itoh, T, Hashimoto, W, Murata, K. | | Deposit date: | 2018-06-29 | | Release date: | 2019-05-15 | | Method: | X-RAY DIFFRACTION (2.609 Å) | | Cite: | Creation of haemoglobin A1c direct oxidase from fructosyl peptide oxidase by combined structure-based site specific mutagenesis and random mutagenesis.
Sci Rep, 9, 2019
|
|
7PKD
 
 | | C-reactive protein decamer at pH 7.5 with phosphocholine ligand | | Descriptor: | C-reactive protein, CALCIUM ION, PHOSPHOCHOLINE | | Authors: | Noone, D.P, Sharp, T.H. | | Deposit date: | 2021-08-25 | | Release date: | 2021-12-22 | | Last modified: | 2022-01-12 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-Electron Microscopy and Biochemical Analysis Offer Insights Into the Effects of Acidic pH, Such as Occur During Acidosis, on the Complement Binding Properties of C-Reactive Protein.
Front Immunol, 12, 2021
|
|
7PKH
 
 | | C-reactive protein decamer at pH 5 with phosphocholine ligand | | Descriptor: | C-reactive protein, CALCIUM ION, PHOSPHOCHOLINE | | Authors: | Noone, D.P, Sharp, T.H. | | Deposit date: | 2021-08-25 | | Release date: | 2021-12-22 | | Last modified: | 2022-01-12 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cryo-Electron Microscopy and Biochemical Analysis Offer Insights Into the Effects of Acidic pH, Such as Occur During Acidosis, on the Complement Binding Properties of C-Reactive Protein.
Front Immunol, 12, 2021
|
|
4QAC
 
 | | X-RAY STRUCTURE OF ACETYLCHOLINE BINDING PROTEIN (ACHBP) IN COMPLEX WITH 4-(4-methylpiperidin-1-yl)-6-(4-(trifluoromethyl)phenyl)pyrimidin-2-amine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(4-methylpiperidin-1-yl)-6-[4-(trifluoromethyl)phenyl]pyrimidin-2-amine, Acetylcholine-binding protein, ... | | Authors: | Kaczanowska, K, Harel, M, Radic, Z, Changeux, J.-P, Finn, M.G, Taylor, P. | | Deposit date: | 2014-05-03 | | Release date: | 2014-07-16 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for cooperative interactions of substituted 2-aminopyrimidines with the acetylcholine binding protein.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
7PKG
 
 | | C-reactive protein pentamer at pH 5 | | Descriptor: | C-reactive protein, CALCIUM ION | | Authors: | Noone, D.P, Sharp, T.H. | | Deposit date: | 2021-08-25 | | Release date: | 2021-12-22 | | Last modified: | 2022-01-12 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-Electron Microscopy and Biochemical Analysis Offer Insights Into the Effects of Acidic pH, Such as Occur During Acidosis, on the Complement Binding Properties of C-Reactive Protein.
Front Immunol, 12, 2021
|
|
6LK6
 
 | | MLKL mutant - T357AS358A | | Descriptor: | Mixed lineage kinase domain-like protein | | Authors: | Wang, H, Li, S, Zhang, Y. | | Deposit date: | 2019-12-18 | | Release date: | 2020-12-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | The MLKL kinase-like domain dimerization is an indispensable step of mammalian MLKL activation in necroptosis signaling.
Cell Death Dis, 12, 2021
|
|
6XRE
 
 | | Structure of the p53/RNA polymerase II assembly | | Descriptor: | Cellular tumor antigen p53, DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11-a, ... | | Authors: | Liou, S.-H, Singh, S, Singer, R.H, Coleman, R.A, Liu, W. | | Deposit date: | 2020-07-12 | | Release date: | 2021-03-24 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structure of the p53/RNA polymerase II assembly.
Commun Biol, 4, 2021
|
|
8CM3
 
 | |
3IL1
 
 | | Crystal structure of the AMPA subunit GluR2 bound to the allosteric modulator, IDRA-21 | | Descriptor: | (3S)-7-chloro-3-methyl-3,4-dihydro-2H-1,2,4-benzothiadiazine 1,1-dioxide, GLUTAMIC ACID, Glutamate receptor 2, ... | | Authors: | Ahmed, A.H, Ptak, C.P, Oswald, R.E. | | Deposit date: | 2009-08-06 | | Release date: | 2009-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | Probing the allosteric modulator binding site of GluR2 with thiazide derivatives
Biochemistry, 48, 2009
|
|
3ILT
 
 | | Crystal structure of the AMPA subunit GluR2 bound to the allosteric modulator, trichlormethiazide | | Descriptor: | 6-CHLORO-3-(DICHLOROMETHYL)-3,4-DIHYDRO-2H-1,2,4-BENZOTHIADIAZINE-7-SULFONAMIDE 1,1-DIOXIDE, GLUTAMIC ACID, Glutamate receptor 2, ... | | Authors: | Ahmed, A.H, Ptak, C.P, Oswald, R.E. | | Deposit date: | 2009-08-07 | | Release date: | 2009-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.107 Å) | | Cite: | Probing the allosteric modulator binding site of GluR2 with thiazide derivatives
Biochemistry, 48, 2009
|
|
6VAP
 
 | | Structure of the type II thioesterase BorB from the borrelidin biosynthetic cluster | | Descriptor: | Thioesterase | | Authors: | Pereira, J.H, Curran, S.C, Baluyot, M.-J, Lake, J, Putz, H, Rosenburg, D, Keasling, J, Adams, P.D. | | Deposit date: | 2019-12-17 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structure and Function of BorB, the Type II Thioesterase from the Borrelidin Biosynthetic Gene Cluster.
Biochemistry, 59, 2020
|
|
4QNC
 
 | | Crystal structure of a SemiSWEET in an occluded state | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, PENTADECANE, chemical transport protein | | Authors: | Yan, X, Yuyong, T, Liang, F, Perry, K. | | Deposit date: | 2014-06-17 | | Release date: | 2014-09-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.388 Å) | | Cite: | Structures of bacterial homologues of SWEET transporters in two distinct conformations.
Nature, 515, 2014
|
|
7PPJ
 
 | | human SLFN5 | | Descriptor: | Schlafen family member 5, ZINC ION | | Authors: | Lammens, K, Metzner, F.J. | | Deposit date: | 2021-09-14 | | Release date: | 2022-01-26 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Structural and biochemical characterization of human Schlafen 5.
Nucleic Acids Res., 50, 2022
|
|
6VBH
 
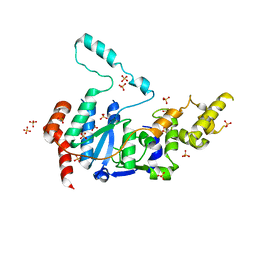 | | Human XPG endonuclease catalytic domain | | Descriptor: | DNA repair protein complementing XP-G cells,Flap endonuclease 1, SULFATE ION | | Authors: | Tsutakawa, S.E, Arvai, A.S, Tainer, J.A. | | Deposit date: | 2019-12-18 | | Release date: | 2020-06-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.995 Å) | | Cite: | Human XPG nuclease structure, assembly, and activities with insights for neurodegeneration and cancer from pathogenic mutations.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6VH7
 
 | | Doublet Tau Fibril from Corticobasal Degeneration Human Brain Tissue | | Descriptor: | Microtubule-associated protein tau | | Authors: | Arakhamia, T, Lee, C.E, Carlomagno, Y, Duong, D.M, Kundinger, S.R, Wang, K, Williams, D, DeTure, M, Dickson, D.W, Cook, C.N, Seyfried, N.T, Petrucelli, L, Fitzpatrick, A.W.P. | | Deposit date: | 2020-01-09 | | Release date: | 2020-03-04 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Posttranslational Modifications Mediate the Structural Diversity of Tauopathy Strains.
Cell, 180, 2020
|
|
7Q3Z
 
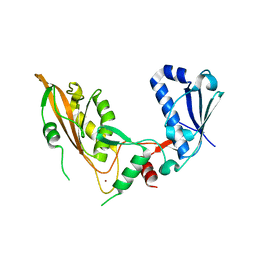 | | DNA/RNA binding protein | | Descriptor: | SODIUM ION, Schlafen family member 5, ZINC ION | | Authors: | Huber, E, Lammens, K. | | Deposit date: | 2021-10-29 | | Release date: | 2022-01-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and biochemical characterization of human Schlafen 5.
Nucleic Acids Res., 50, 2022
|
|
6M71
 
 | | SARS-Cov-2 RNA-dependent RNA polymerase in complex with cofactors | | Descriptor: | Non-structural protein 7, Non-structural protein 8, RNA-directed RNA polymerase | | Authors: | Gao, Y, Yan, L, Huang, Y, Liu, F, Cao, L, Wang, T, Wang, Q, Lou, Z, Rao, Z. | | Deposit date: | 2020-03-16 | | Release date: | 2020-04-01 | | Last modified: | 2021-03-10 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure of the RNA-dependent RNA polymerase from COVID-19 virus.
Science, 368, 2020
|
|
5IJS
 
 | | Crystal structure of autotaxin with orthovanadate bound as a trigonal bipyramidal intermediate analog | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 7alpha-hydroxycholesterol, CALCIUM ION, ... | | Authors: | Hausmann, J, Joosten, R.P, Perrakis, A. | | Deposit date: | 2016-03-02 | | Release date: | 2016-06-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural snapshots of the catalytic cycle of the phosphodiesterase Autotaxin.
J.Struct.Biol., 195, 2016
|
|
