8B6K
 
 | |
8B67
 
 | |
8B79
 
 | |
8B76
 
 | |
2HOJ
 
 | |
6AR5
 
 | |
6Q4U
 
 | | KlenTaq DNA pol in a closed ternary complex with 7-deaza-7-(2-(2-hydroxyethoxy)-N-(prop-2-yn-1-yl)acetamide)-2-dATP | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*AP*AP*CP*TP*GP*TP*GP*GP*CP*CP*GP*TP*GP*GP*TP*C)-3'), DNA (5'-D(*GP*AP*CP*CP*AP*CP*GP*GP*CP*CP*AP*(DOC))-3'), ... | | Authors: | Kropp, H.M, Diederichs, K, Marx, A. | | Deposit date: | 2018-12-06 | | Release date: | 2019-02-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.005 Å) | | Cite: | The Structure of an Archaeal B-Family DNA Polymerase in Complex with a Chemically Modified Nucleotide.
Angew.Chem.Int.Ed.Engl., 58, 2019
|
|
4P36
 
 | | Crystal structure of DJ-1 With Zn(II) bound (crystal 2) | | Descriptor: | 2,5,8,11,14,17-HEXAOXANONADECAN-19-OL, Protein DJ-1, ZINC ION | | Authors: | Tashiro, S, Wu, C.-X, Hoang, Q.Q, Caaveiro, J.M.M, Tsumoto, K. | | Deposit date: | 2014-03-05 | | Release date: | 2014-04-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.182 Å) | | Cite: | Thermodynamic and Structural Characterization of the Specific Binding of Zn(II) to Human Protein DJ-1.
Biochemistry, 53, 2014
|
|
4P35
 
 | | Crystal structure of DJ-1 with Zinc(II) bound (crystal I) | | Descriptor: | CHLORIDE ION, GLYCEROL, Protein DJ-1, ... | | Authors: | Tashiro, S, Wu, C.-X, Hoang, Q.Q, Caaveiro, J.M.M, Tsumoto, K. | | Deposit date: | 2014-03-05 | | Release date: | 2014-04-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Thermodynamic and Structural Characterization of the Specific Binding of Zn(II) to Human Protein DJ-1.
Biochemistry, 53, 2014
|
|
6WC5
 
 | | Crystal Structure of a Ternary MEF2B/NKX2-5/myocardin enhancer DNA Complex | | Descriptor: | Homeobox protein Nkx-2.5, Myocardin enhancer DNA, Myocyte-specific enhancer factor 2B | | Authors: | Chen, L, Lei, X. | | Deposit date: | 2020-03-29 | | Release date: | 2020-07-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structures of Ternary Complexes of MEF2 and NKX2-5 Bound to DNA Reveal a Disease Related Protein-Protein Interaction Interface.
J.Mol.Biol., 432, 2020
|
|
3ABD
 
 | | Structure of human REV7 in complex with a human REV3 fragment in a monoclinic crystal | | Descriptor: | DNA polymerase zeta catalytic subunit, Mitotic spindle assembly checkpoint protein MAD2B | | Authors: | Hara, K, Hashimoto, H, Murakumo, Y, Kobayashi, S, Kogame, T, Unzai, S, Akashi, S, Takeda, S, Shimizu, T, Sato, M. | | Deposit date: | 2009-12-07 | | Release date: | 2010-02-16 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of human REV7 in complex with a human REV3 fragment and structural implication of the interaction between DNA polymerase {zeta} and REV1
J.Biol.Chem., 285, 2010
|
|
3ABE
 
 | | Structure of human REV7 in complex with a human REV3 fragment in a tetragonal crystal | | Descriptor: | DNA polymerase zeta catalytic subunit, Mitotic spindle assembly checkpoint protein MAD2B | | Authors: | Hara, K, Hashimoto, H, Murakumo, Y, Kobayashi, S, Kogame, T, Unzai, S, Akashi, S, Takeda, S, Shimizu, T, Sato, M. | | Deposit date: | 2009-12-07 | | Release date: | 2010-02-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of human REV7 in complex with a human REV3 fragment and structural implication of the interaction between DNA polymerase {zeta} and REV1
J.Biol.Chem., 285, 2010
|
|
6ZY7
 
 | | Cryo-EM structure of the entire Human topoisomerase II alpha in State 1 | | Descriptor: | (5S,5aR,8aR,9R)-9-(4-hydroxy-3,5-dimethoxyphenyl)-8-oxo-5,5a,6,8,8a,9-hexahydrofuro[3',4':6,7]naphtho[2,3-d][1,3]dioxol -5-yl 4,6-O-[(1R)-ethylidene]-beta-D-glucopyranoside, DNA (5'-D(*CP*GP*CP*GP*CP*AP*TP*CP*GP*TP*CP*AP*TP*CP*CP*TP*C)-3'), DNA (5'-D(*GP*AP*GP*GP*AP*TP*GP*AP*CP*GP*AP*TP*G)-3'), ... | | Authors: | Vanden Broeck, A, Lamour, V. | | Deposit date: | 2020-07-30 | | Release date: | 2021-05-26 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (4.64 Å) | | Cite: | Structural basis for allosteric regulation of Human Topoisomerase II alpha.
Nat Commun, 12, 2021
|
|
6ZY6
 
 | | Cryo-EM structure of the Human topoisomerase II alpha DNA-binding/cleavage domain in State 2 | | Descriptor: | (5S,5aR,8aR,9R)-9-(4-hydroxy-3,5-dimethoxyphenyl)-8-oxo-5,5a,6,8,8a,9-hexahydrofuro[3',4':6,7]naphtho[2,3-d][1,3]dioxol -5-yl 4,6-O-[(1R)-ethylidene]-beta-D-glucopyranoside, DNA (5'-D(*CP*GP*CP*GP*CP*AP*TP*CP*GP*TP*CP*AP*TP*CP*CP*TP*C)-3'), DNA (5'-D(*GP*AP*GP*GP*AP*TP*GP*AP*CP*GP*AP*TP*G)-3'), ... | | Authors: | Vanden Broeck, A, Lamour, V. | | Deposit date: | 2020-07-30 | | Release date: | 2021-05-26 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural basis for allosteric regulation of Human Topoisomerase II alpha.
Nat Commun, 12, 2021
|
|
6ZY8
 
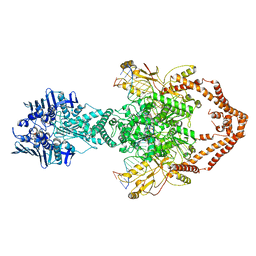 | | Cryo-EM structure of the entire Human topoisomerase II alpha in State 2 | | Descriptor: | (5S,5aR,8aR,9R)-9-(4-hydroxy-3,5-dimethoxyphenyl)-8-oxo-5,5a,6,8,8a,9-hexahydrofuro[3',4':6,7]naphtho[2,3-d][1,3]dioxol -5-yl 4,6-O-[(1R)-ethylidene]-beta-D-glucopyranoside, DNA (5'-D(*CP*GP*CP*GP*CP*AP*TP*CP*GP*TP*CP*AP*TP*CP*CP*TP*C)-3'), DNA (5'-D(*GP*AP*GP*GP*AP*TP*GP*AP*CP*GP*AP*TP*G)-3'), ... | | Authors: | Vanden Broeck, A, Lamour, V. | | Deposit date: | 2020-07-30 | | Release date: | 2021-05-26 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (7.4 Å) | | Cite: | Structural basis for allosteric regulation of Human Topoisomerase II alpha.
Nat Commun, 12, 2021
|
|
6ZY5
 
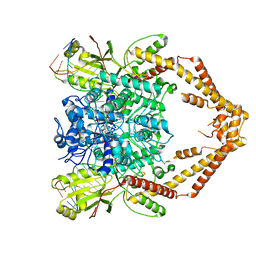 | | Cryo-EM structure of the Human topoisomerase II alpha DNA-binding/cleavage domain in State 1 | | Descriptor: | (5S,5aR,8aR,9R)-9-(4-hydroxy-3,5-dimethoxyphenyl)-8-oxo-5,5a,6,8,8a,9-hexahydrofuro[3',4':6,7]naphtho[2,3-d][1,3]dioxol -5-yl 4,6-O-[(1R)-ethylidene]-beta-D-glucopyranoside, DNA (5'-D(*CP*GP*CP*GP*CP*AP*TP*CP*GP*TP*CP*AP*TP*CP*CP*TP*C)-3'), DNA (5'-D(*GP*AP*GP*GP*AP*TP*GP*AP*CP*GP*AP*TP*G)-3'), ... | | Authors: | Vanden Broeck, A, Lamour, V. | | Deposit date: | 2020-07-30 | | Release date: | 2021-05-26 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis for allosteric regulation of Human Topoisomerase II alpha.
Nat Commun, 12, 2021
|
|
7TO2
 
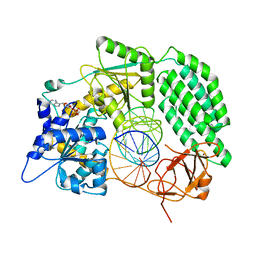 | |
4TUS
 
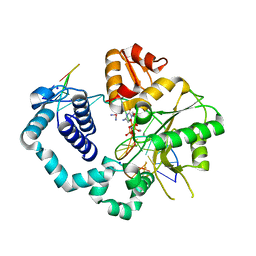 | |
1NOY
 
 | | DNA POLYMERASE (E.C.2.7.7.7)/DNA COMPLEX | | Descriptor: | DNA (5'-D(*TP*TP*T)-3'), MANGANESE (II) ION, PROTEIN (DNA POLYMERASE (E.C.2.7.7.7)), ... | | Authors: | Wang, J, Yu, P, Lin, T.C, Konigsberg, W.H, Steitz, T.A. | | Deposit date: | 1996-02-16 | | Release date: | 1996-10-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of an NH2-terminal fragment of T4 DNA polymerase and its complexes with single-stranded DNA and with divalent metal ions.
Biochemistry, 35, 1996
|
|
6AOC
 
 | | Crystal Structure of an N-Hydroxythienopyrimidine-2,4-dione RNase H Active Site Inhibitor with Multiple Binding Modes to HIV Reverse Transcriptase | | Descriptor: | 1,2-ETHANEDIOL, 6-benzyl-3-hydroxythieno[2,3-d]pyrimidine-2,4(1H,3H)-dione, MANGANESE (II) ION, ... | | Authors: | Kirby, K.A, Sarafianos, S.G. | | Deposit date: | 2017-08-15 | | Release date: | 2018-08-08 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Design, synthesis and biological evaluations of N-Hydroxy thienopyrimidine-2,4-diones as inhibitors of HIV reverse transcriptase-associated RNase H.
Eur J Med Chem, 141, 2017
|
|
5XPO
 
 | | Crystal structure of VDR-LBD complexed with 25-(hydroxyphenyl)-2-methylidene-19,26,27-trinor-25-oxo-1-hydroxyvitamin D3 | | Descriptor: | (5~{R})-5-[(1~{R},3~{a}~{S},4~{E},7~{a}~{R})-7~{a}-methyl-4-[2-[(3~{R},5~{R})-4-methylidene-3,5-bis(oxidanyl)cyclohexyl idene]ethylidene]-2,3,3~{a},5,6,7-hexahydro-1~{H}-inden-1-yl]-1-(4-hydroxyphenyl)hexan-1-one, Mediator of RNA polymerase II transcription subunit 1, Vitamin D3 receptor | | Authors: | Kato, A, Itoh, T, Yamamoto, K. | | Deposit date: | 2017-06-03 | | Release date: | 2018-06-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Vitamin D Analogues with a p-Hydroxyphenyl Group at the C25 Position: Crystal Structure of Vitamin D Receptor Ligand-Binding Domain Complexed with the Ligand Explains the Mechanism Underlying Full Antagonistic Action
J. Med. Chem., 60, 2017
|
|
5XPN
 
 | | Crystal structure of VDR-LBD complexed with 25RS-(hydroxyphenyl)-25-methoxy-2-methylidene-19,26,27-trinor-1-hydroxyvitamin D3 | | Descriptor: | (1~{R},3~{R})-5-[(2~{E})-2-[(1~{R},3~{a}~{S},7~{a}~{R})-1-[(2~{R},6~{R})-6-(4-hydroxyphenyl)-6-methoxy-hexan-2-yl]-7~{a}-methyl-2,3,3~{a},5,6,7-hexahydro-1~{H}-inden-4-ylidene]ethylidene]-2-methylidene-cyclohexane-1,3-diol, (1~{R},3~{R})-5-[(2~{E})-2-[(1~{R},3~{a}~{S},7~{a}~{R})-1-[(2~{R},6~{S})-6-(4-hydroxyphenyl)-6-methoxy-hexan-2-yl]-7~{a}-methyl-2,3,3~{a},5,6,7-hexahydro-1~{H}-inden-4-ylidene]ethylidene]-2-methylidene-cyclohexane-1,3-diol, Mediator of RNA polymerase II transcription subunit 1, ... | | Authors: | Kato, A, Itoh, T, Yamamoto, K. | | Deposit date: | 2017-06-03 | | Release date: | 2018-07-11 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Vitamin D Analogues with a p-Hydroxyphenyl Group at the C25 Position: Crystal Structure of Vitamin D Receptor Ligand-Binding Domain Complexed with the Ligand Explains the Mechanism Underlying Full Antagonistic Action
J. Med. Chem., 60, 2017
|
|
5XPP
 
 | | Crystal structure of VDR-LBD complexed with 25RS-(Hydroxyphenyl)-2-methylidene-19,26,27-trinor-1,25-dihydroxyvitamin D3 | | Descriptor: | (1~{R},3~{R})-5-[(2~{E})-2-[(1~{R},3~{a}~{S},7~{a}~{R})-1-[(2~{R},6~{R})-6-(4-hydroxyphenyl)-6-oxidanyl-hexan-2-yl]-7~{ a}-methyl-2,3,3~{a},5,6,7-hexahydro-1~{H}-inden-4-ylidene]ethylidene]-2-methylidene-cyclohexane-1,3-diol, Mediator of RNA polymerase II transcription subunit 1, Vitamin D3 receptor | | Authors: | Kato, A, Itoh, T, Yamamoto, K. | | Deposit date: | 2017-06-03 | | Release date: | 2018-06-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Vitamin D Analogues with a p-Hydroxyphenyl Group at the C25 Position: Crystal Structure of Vitamin D Receptor Ligand-Binding Domain Complexed with the Ligand Explains the Mechanism Underlying Full Antagonistic Action
J. Med. Chem., 60, 2017
|
|
5TBB
 
 | |
5TB9
 
 | |
