8BEH
 
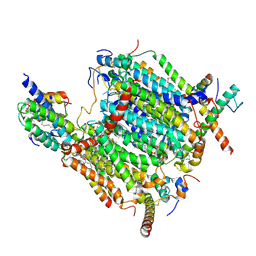 | | Cryo-EM structure of the Arabidopsis thaliana I+III2 supercomplex (CI membrane tip) | | Descriptor: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, (7S)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, ... | | Authors: | Klusch, N, Kuehlbrandt, W. | | Deposit date: | 2022-10-21 | | Release date: | 2023-01-11 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.29 Å) | | Cite: | Cryo-EM structure of the respiratory I + III 2 supercomplex from Arabidopsis thaliana at 2 angstrom resolution.
Nat.Plants, 9, 2023
|
|
1UGX
 
 | | Crystal structure of jacalin- Me-alpha-T-antigen (Gal-beta(1-3)-GalNAc-alpha-o-Me) complex | | Descriptor: | Agglutinin alpha chain, Agglutinin beta-3 chain, beta-D-galactopyranose-(1-3)-methyl 2-acetamido-2-deoxy-alpha-D-galactopyranoside | | Authors: | Jeyaprakash, A.A, Katiyar, S, Swaminathan, C.P, Sekar, K, Surolia, A, Vijayan, M. | | Deposit date: | 2003-06-22 | | Release date: | 2003-09-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Basis of the Carbohydrate Specificities of Jacalin: An X-ray and Modeling Study
J.MOL.BIOL., 332, 2003
|
|
8B90
 
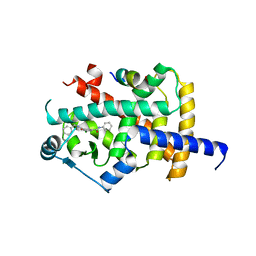 | | Crystal structure of PPARG and NCOR2 with an inverse agonist (compound 7d) | | Descriptor: | 5-chloranyl-~{N}3-phenyl-~{N}1-(phenylmethyl)benzene-1,3-dicarboxamide, Nuclear receptor corepressor 2, Peroxisome proliferator-activated receptor gamma | | Authors: | Friberg, A, Orsi, D.L, Pook, E, Siegel, S, Lemke, C.T, Stellfeld, T, Puetter, V, Goldstein, J. | | Deposit date: | 2022-10-05 | | Release date: | 2022-12-28 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery and characterization of orally bioavailable 4-chloro-6-fluoroisophthalamides as covalent PPARG inverse-agonists.
Bioorg.Med.Chem., 78, 2022
|
|
1U5C
 
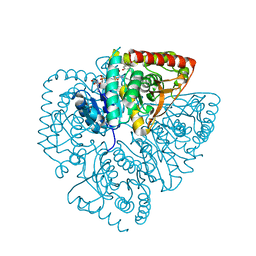 | | Plasmodium falciparum lactate dehydrogenase complexed with 3,7-dihydroxynaphthalene-2-carboxylic acid and NAD+ | | Descriptor: | 3,7-DIHYDROXY-2-NAPHTHOIC ACID, L-lactate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Conners, R, Cameron, A, Read, J, Schambach, F, Sessions, R.B, Brady, R.L. | | Deposit date: | 2004-07-27 | | Release date: | 2005-06-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Mapping the binding site for gossypol-like inhibitors of Plasmodium falciparum lactate dehydrogenase.
Mol.Biochem.Parasitol., 142, 2005
|
|
1U5R
 
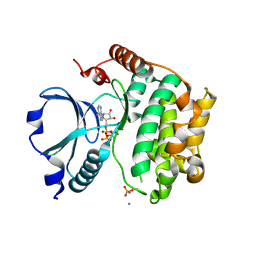 | | Crystal Structure of the TAO2 Kinase Domain: Activation and Specifity of a Ste20p MAP3K | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Zhou, T, Raman, M, Gao, Y, Earnest, S, Chen, Z, Machius, M, Cobb, M.H, Goldsmith, E.J. | | Deposit date: | 2004-07-28 | | Release date: | 2004-10-12 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of the TAO2 Kinase Domain; Activation and Specificity of a Ste20p MAP3K.
Structure, 12, 2004
|
|
1U68
 
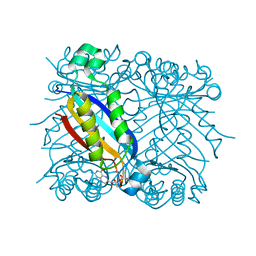 | | DHNA 7,8 DIHYDRONEOPTERIN COMPLEX | | Descriptor: | 2-AMINO-7,8-DIHYDRO-6-(1,2,3-TRIHYDROXYPROPYL)-4(1H)-PTERIDINONE, Dihydroneopterin aldolase | | Authors: | Sanders, W.J, Nienaber, V.L, Lerner, C.G, McCall, J.O, Merrick, S.M, Swanson, S.J, Harlan, J.E, Stoll, V.S, Stamper, G.F, Betz, S.F, Condroski, K.R, Meadows, R.P, Severin, J.M, Walter, K.A, Magdalinos, P, Jakob, C.G, Wagner, R, Beutel, B.A. | | Deposit date: | 2004-07-29 | | Release date: | 2004-10-19 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery of potent inhibitors of dihydroneopterin aldolase using CrystaLEAD high-throughput X-ray crystallographic screening and structure-directed lead optimization.
J.MED.CHEM., 47, 2004
|
|
8AVV
 
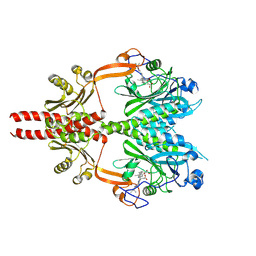 | | Cryo-EM structure of DrBphP photosensory module in Pr state | | Descriptor: | 3-[2-[(Z)-[3-(2-carboxyethyl)-5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-pyrrol-1-ium -2-ylidene]methyl]-5-[(Z)-[(3E)-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-4-methyl-1H-pyrrol-3- yl]propanoic acid, Bacteriophytochrome,Response regulator | | Authors: | Wahlgren, W.Y, Takala, H, Westenhoff, S. | | Deposit date: | 2022-08-27 | | Release date: | 2022-12-21 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural mechanism of signal transduction in a phytochrome histidine kinase.
Nat Commun, 13, 2022
|
|
1U6E
 
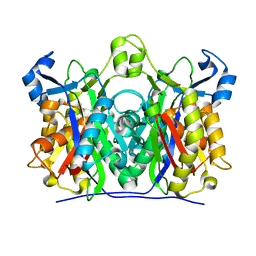 | | 1.85 Angstrom Crystal Structure of the C112A Mutant of Mycobacterium Tuberculosis Beta-Ketoacyl-Acyl Carrier Protein Synthase III (FabH) | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] synthase III, CHLORIDE ION | | Authors: | Mussayev, F, Sachedeva, S, Scarsdale, J.N, Reynolds, K.A, Wright, H.T. | | Deposit date: | 2004-07-29 | | Release date: | 2005-05-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of a substrate complex of Mycobacterium tuberculosis beta-ketoacyl-acyl carrier protein synthase III (FabH) with lauroyl-coenzyme A.
J.Mol.Biol., 346, 2005
|
|
8BBG
 
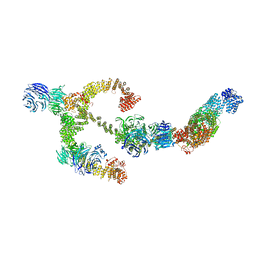 | | Structure of the IFT-A complex; anterograde IFT-A train model | | Descriptor: | Intraflagellar transport protein 122 homolog, Intraflagellar transport protein 140 homolog, Intraflagellar transport protein 43 homolog, ... | | Authors: | Hesketh, S.J, Mukhopadhyay, A.G, Nakamura, D, Toropova, K, Roberts, A.J. | | Deposit date: | 2022-10-12 | | Release date: | 2022-12-21 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | IFT-A structure reveals carriages for membrane protein transport into cilia.
Cell, 185, 2022
|
|
8BAH
 
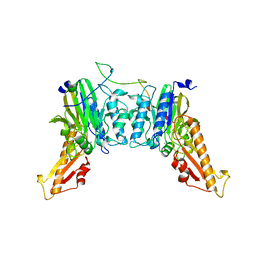 | | Human Mre11-Nbs1 complex | | Descriptor: | Double-strand break repair protein MRE11, MANGANESE (II) ION, Nibrin | | Authors: | Bartho, J.D, Rotheneder, M, Stakyte, K, Lammens, K, Hopfner, K.P. | | Deposit date: | 2022-10-11 | | Release date: | 2023-01-11 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (4.13 Å) | | Cite: | Cryo-EM structure of the Mre11-Rad50-Nbs1 complex reveals the molecular mechanism of scaffolding functions.
Mol.Cell, 83, 2023
|
|
1U6Z
 
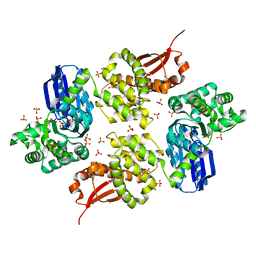 | |
1UH2
 
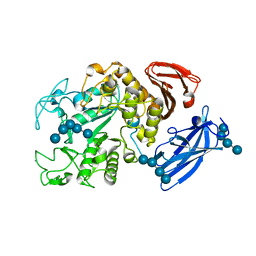 | | Thermoactinomyces vulgaris R-47 alpha-amylase/malto-hexaose complex | | Descriptor: | CALCIUM ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Abe, A, Tonozuka, T, Sakano, Y, Kamitori, S. | | Deposit date: | 2003-06-23 | | Release date: | 2004-01-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Complex Structures of Thermoactinomyces vulgaris R-47 alpha-Amylase 1 with Malto-oligosaccharides Demonstrate the Role of Domain N Acting as a Starch-binding Domain
J.Mol.Biol., 335, 2004
|
|
8B0S
 
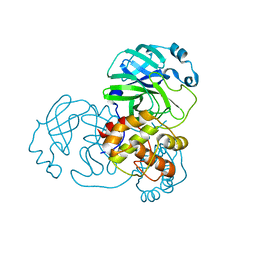 | | SARS-COV-2 Main Protease adduct with Au(NHC)Cl | | Descriptor: | 3C-like proteinase nsp5, GOLD ION | | Authors: | Massai, L, Grifagni, D, Desantis, A, Geri, A, Calderone, V, Cantini, F, Messori, L, Banci, L. | | Deposit date: | 2022-09-08 | | Release date: | 2022-12-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Gold-Based Metal Drugs as Inhibitors of Coronavirus Proteins: The Inhibition of SARS-CoV-2 Main Protease by Auranofin and Its Analogs.
Biomolecules, 12, 2022
|
|
8B8Z
 
 | | Crystal structure of mutant PPARG (C313A) and NCOR2 with an inverse agonist (compound 7e) | | Descriptor: | 4,5-bis(chloranyl)-~{N}3-phenyl-~{N}1-(phenylmethyl)benzene-1,3-dicarboxamide, Nuclear receptor corepressor 2, Peroxisome proliferator-activated receptor gamma | | Authors: | Friberg, A, Orsi, D.L, Pook, E, Siegel, S, Lemke, C.T, Stellfeld, T, Puetter, V, Goldstein, J. | | Deposit date: | 2022-10-05 | | Release date: | 2022-12-28 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Discovery and characterization of orally bioavailable 4-chloro-6-fluoroisophthalamides as covalent PPARG inverse-agonists.
Bioorg.Med.Chem., 78, 2022
|
|
1UEY
 
 | | Solution Structure of The First Fibronectin Type III Domain of Human KIAA0343 protein | | Descriptor: | KIAA0343 protein | | Authors: | Zhao, C, Kigawa, T, Muto, Y, Koshiba, S, Tochio, N, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-05-22 | | Release date: | 2003-11-22 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of The First Fibronectin Type III Domain of Human KIAA0343 protein
To be Published
|
|
1UFY
 
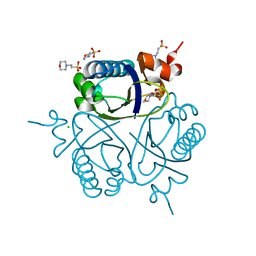 | |
8BFJ
 
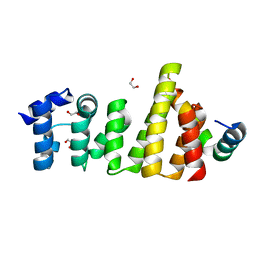 | | CNOT11-GGNBP2 complex | | Descriptor: | 1,2-ETHANEDIOL, CCR4-NOT transcription complex subunit 11, Gametogenetin-binding protein 2 | | Authors: | Basquin, J, Ozgur, S, Conti, E. | | Deposit date: | 2022-10-26 | | Release date: | 2022-12-28 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | The human CNOT1-CNOT10-CNOT11 complex forms a structural platform for protein-protein interactions.
Cell Rep, 42, 2023
|
|
8BDP
 
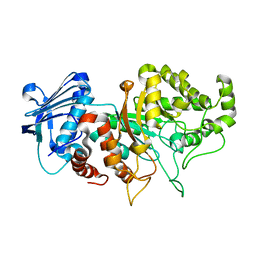 | |
1UG2
 
 | | Solution Structure of Mouse Hypothetical Gene (2610100B20Rik) Product Homologous to Myb DNA-binding Domain | | Descriptor: | 2610100B20Rik gene product | | Authors: | Zhao, C, Kigawa, T, Tochio, N, Koshiba, S, Inoue, M, Shirouzu, M, Terada, T, Yabuki, T, Aoki, M, Seki, E, Matsuda, T, Tanaka, A, Osanai, T, Arakawa, T, Carninci, P, Kawai, J, Hayashizaki, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-06-11 | | Release date: | 2004-06-22 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Mouse Hypothetical Gene (2610100B20Rik) Product Homologous to Myb DNA-binding Domain
To be Published
|
|
1U7I
 
 | | Crystal Structure of Protein of Unknown Function PA1358 from Pseudomonas aeruginosa | | Descriptor: | hypothetical protein | | Authors: | Osipiuk, J, Xu, X, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-08-03 | | Release date: | 2004-08-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | X-ray crystal structure of hypothetical protein PA1358 from Pseudomonas aeruginosa
To be Published
|
|
8BEF
 
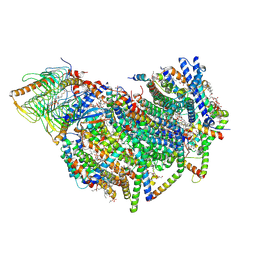 | | Cryo-EM structure of the Arabidopsis thaliana I+III2 supercomplex (CI membrane core) | | Descriptor: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, (7S)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, ... | | Authors: | Klusch, N, Kuehlbrandt, W. | | Deposit date: | 2022-10-21 | | Release date: | 2023-01-11 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.13 Å) | | Cite: | Cryo-EM structure of the respiratory I + III 2 supercomplex from Arabidopsis thaliana at 2 angstrom resolution.
Nat.Plants, 9, 2023
|
|
1UGQ
 
 | | Crystal structure of apoenzyme of Co-type nitrile hydratase | | Descriptor: | Nitrile Hydratase alpha subunit, Nitrile Hydratase beta subunit | | Authors: | Miyanaga, A, Fushinobu, S, Ito, K, Shoun, H, Wakagi, T. | | Deposit date: | 2003-06-17 | | Release date: | 2004-06-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mutational and structural analysis of cobalt-containing nitrile hydratase on substrate and metal binding
Eur.J.Biochem., 271, 2004
|
|
8BGO
 
 | | N,N-diacetylchitobiose deacetylase from Pyrococcus chitonophagus with substrate N,N-diacetylchitobiose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Diacetylchitobiose deacetylase, ZINC ION | | Authors: | Rypniewski, W, Bejger, M, Biniek-Antosiak, K. | | Deposit date: | 2022-10-28 | | Release date: | 2023-01-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.08 Å) | | Cite: | Structural, Thermodynamic and Enzymatic Characterization of N , N -Diacetylchitobiose Deacetylase from Pyrococcus chitonophagus.
Int J Mol Sci, 23, 2022
|
|
1UH4
 
 | | Thermoactinomyces vulgaris R-47 alpha-amylase 1/malto-tridecaose complex | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, alpha-D-glucopyranose, ... | | Authors: | Abe, A, Tonozuka, T, Sakano, Y, Kamitori, S. | | Deposit date: | 2003-06-24 | | Release date: | 2004-01-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Complex Structures of Thermoactinomyces vulgaris R-47 alpha-Amylase 1 with Malto-oligosaccharides Demonstrate the Role of Domain N Acting as a Starch-binding Domain
J.Mol.Biol., 335, 2004
|
|
8AYH
 
 | | Structure of Complement C5 in Complex with small molecule inhibitor and CVF | | Descriptor: | 5-methoxy-2-[[(1~{S})-1-(2-methoxyphenyl)ethyl]carbamoylamino]-4-(4-methylpentoxy)benzoic acid, Cobra venom factor, Complement C5 alpha chain, ... | | Authors: | Srinivas, H. | | Deposit date: | 2022-09-02 | | Release date: | 2022-12-21 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | A small-molecule inhibitor of C5 complement protein.
Nat.Chem.Biol., 15, 2019
|
|
