6OOX
 
 | |
6OF5
 
 | | The crystal structure of dodecyloxy(naphthalen-1-yl)methylphosphonic acid in complex with red kidney bean purple acid phosphatase | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, FE (III) ION, ... | | Authors: | Feder, D, Schenk, G, Guddat, L.W, Hussein, W.M, McGeary, R.P. | | Deposit date: | 2019-03-28 | | Release date: | 2019-09-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Synthesis, evaluation and structural investigations of potent purple acid phosphatase inhibitors as drug leads for osteoporosis.
Eur.J.Med.Chem., 182, 2019
|
|
1MYM
 
 | |
4G9B
 
 | | Crystal structure of beta-phosphoglucomutase homolog from escherichia coli, target efi-501172, with bound mg, open lid | | Descriptor: | Beta-phosphoglucomutase, CHLORIDE ION, GLYCEROL, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Dunaway-Mariano, D, Allen, K.N, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-07-23 | | Release date: | 2012-08-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of beta-phosphoglucomutase homolog from escherichia coli, target efi-501172, with bound mg, open lid
To be Published
|
|
1QHU
 
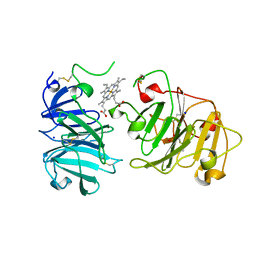 | | MAMMALIAN BLOOD SERUM HAEMOPEXIN DEGLYCOSYLATED AND IN COMPLEX WITH ITS LIGAND HAEM | | Descriptor: | CHLORIDE ION, PHOSPHATE ION, PROTEIN (HEMOPEXIN), ... | | Authors: | Paoli, M, Baker, H.M, Morgan, W.T, Smith, A, Baker, E.N. | | Deposit date: | 1999-05-27 | | Release date: | 1999-10-06 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of hemopexin reveals a novel high-affinity heme site formed between two beta-propeller domains.
Nat.Struct.Biol., 6, 1999
|
|
4XI3
 
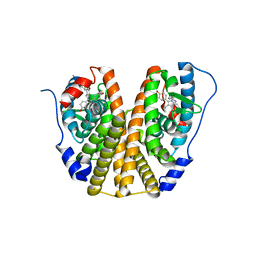 | | Estrogen Receptor Alpha Ligand Binding Domain in Complex with Bazedoxifene | | Descriptor: | Bazedoxifene, Estrogen receptor | | Authors: | Fanning, S.W, Mayne, C.G, Toy, W, Carlson, K, Greene, B, Nowak, J, Walter, R, Panchamukhi, S, Tajhorshid, E, Nettles, K.W, Chandarlapaty, S, Katzenellenbogen, J, Greene, G.L. | | Deposit date: | 2015-01-06 | | Release date: | 2016-01-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.491 Å) | | Cite: | The SERM/SERD bazedoxifene disrupts ESR1 helix 12 to overcome acquired hormone resistance in breast cancer cells.
Elife, 7, 2018
|
|
4TUV
 
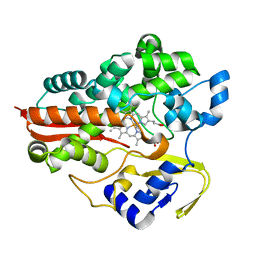 | |
8T2R
 
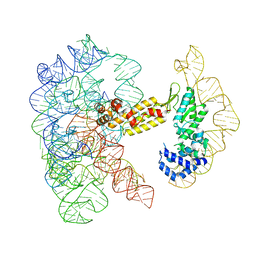 | | Structure of a group II intron ribonucleoprotein in the pre-ligation (pre-2F) state | | Descriptor: | 5'exon, AMMONIUM ION, CALCIUM ION, ... | | Authors: | Xu, L, Liu, T, Chung, K, Pyle, A.M. | | Deposit date: | 2023-06-06 | | Release date: | 2023-11-22 | | Last modified: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into intron catalysis and dynamics during splicing.
Nature, 624, 2023
|
|
1IUZ
 
 | | PLASTOCYANIN | | Descriptor: | COPPER (II) ION, PLASTOCYANIN, SULFATE ION | | Authors: | Shibata, N. | | Deposit date: | 1996-10-06 | | Release date: | 1997-08-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Novel insight into the copper-ligand geometry in the crystal structure of Ulva pertusa plastocyanin at 1.6-A resolution. Structural basis for regulation of the copper site by residue 88.
J.Biol.Chem., 274, 1999
|
|
3H3H
 
 | |
8T2T
 
 | | Structure of a group II intron ribonucleoprotein in the post-ligation (post-2F) state | | Descriptor: | AMMONIUM ION, Group II intron reverse transcriptase/maturase, MAGNESIUM ION, ... | | Authors: | Xu, L, Liu, T, Chung, K, Pyle, A.M. | | Deposit date: | 2023-06-06 | | Release date: | 2023-11-22 | | Last modified: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into intron catalysis and dynamics during splicing.
Nature, 624, 2023
|
|
8BZO
 
 | | Cryo-EM structure of CDK2-CyclinA in complex with p27 from the SCFSKP2 E3 ligase Complex | | Descriptor: | Cyclin-A2, Cyclin-dependent kinase 2, Cyclin-dependent kinase inhibitor 1B | | Authors: | Rowland, R.J, Salamina, M, Endicott, J.A, Noble, M.E. | | Deposit date: | 2022-12-15 | | Release date: | 2023-06-28 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structure of SKP1-SKP2-CKS1 in complex with CDK2-cyclin A-p27KIP1.
Sci Rep, 13, 2023
|
|
6J8V
 
 | | Structure of MOEN5-SSO7D fusion protein in complex with ligand 2 | | Descriptor: | FARNESYL, MoeN5,DNA-binding protein 7d | | Authors: | Ko, T.P, Zhang, L.L, Chen, C.C, Guo, R.T. | | Deposit date: | 2019-01-21 | | Release date: | 2019-04-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Complex structures of MoeN5 with substrate analogues suggest sequential catalytic mechanism.
Biochem. Biophys. Res. Commun., 511, 2019
|
|
5C9J
 
 | | Human CD1c with ligands in A' and F' channel | | Descriptor: | Beta-2-microglobulin, GLYCEROL, LAURIC ACID, ... | | Authors: | Tews, I, Machelett, M.M, Mansour, S, Gadola, S.D. | | Deposit date: | 2015-06-27 | | Release date: | 2016-03-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Cholesteryl esters stabilize human CD1c conformations for recognition by self-reactive T cells.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
1PPV
 
 | |
4Y1Q
 
 | | Crystal Structure of HasA mutant Y75A monomer from Yersinia pseudotuberculosis | | Descriptor: | Extracellular heme acquisition hemophore HasA, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Nagano, S, Hino, T, Kanadani, M, Ozaki, S, Sato, T. | | Deposit date: | 2015-02-08 | | Release date: | 2015-08-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The crystal structure of heme acquisition system A from Yersinia pseudotuberculosis (HasAypt): Roles of the axial ligand Tyr75 and two distal arginines in heme binding
J.Inorg.Biochem., 151, 2015
|
|
1D8F
 
 | | CRYSTAL STRUCTURE OF MMP3 COMPLEXED WITH A PIPERAZINE BASED INHIBITOR. | | Descriptor: | CALCIUM ION, N-HYDROXY-1-(4-METHOXYPHENYL)SULFONYL-4-BENZYLOXYCARBONYL-PIPERAZINE-2-CARBOXAMIDE, STROMELYSIN-1 PRECURSOR, ... | | Authors: | Cheng, M.Y, De, B, Pikul, S, Almstead, N.G, Natchus, M.G, Anastasio, M.V, McPhail, S.J, Snider, C.E, Taiwo, Y.O, Chen, L.Y. | | Deposit date: | 1999-10-22 | | Release date: | 2000-10-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Design and synthesis of piperazine-based matrix metalloproteinase inhibitors.
J.Med.Chem., 43, 2000
|
|
6PIT
 
 | | Estrogen Receptor Alpha Ligand Binding Domain Y537S Mutant in Complex with SRC2 Stapled Peptide 41A and Estradiol | | Descriptor: | ESTRADIOL, Estrogen receptor, Stapled Peptide 41A | | Authors: | Fanning, S.W, Montgomery, J.E, Greene, G.L, Moellering, R.E. | | Deposit date: | 2019-06-27 | | Release date: | 2019-10-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Versatile Peptide Macrocyclization with Diels-Alder Cycloadditions.
J.Am.Chem.Soc., 141, 2019
|
|
7NTF
 
 | | Cryo-EM structure of unliganded O-GlcNAc transferase | | Descriptor: | Isoform 1 of UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunit | | Authors: | Meek, R.W, Blaza, J.N, Davies, G.J. | | Deposit date: | 2021-03-09 | | Release date: | 2021-11-17 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (5.32 Å) | | Cite: | Cryo-EM structure provides insights into the dimer arrangement of the O-linked beta-N-acetylglucosamine transferase OGT.
Nat Commun, 12, 2021
|
|
6PL2
 
 | | TRK-A IN COMPLEX WITH LIGAND 1a | | Descriptor: | High affinity nerve growth factor receptor, N-(3-tert-butyl-1-phenyl-1H-pyrazol-5-yl)-2-{[1-(4-hydroxyphenyl)-1H-tetrazol-5-yl]sulfanyl}acetamide | | Authors: | Subramanian, G. | | Deposit date: | 2019-06-30 | | Release date: | 2019-09-04 | | Last modified: | 2019-09-25 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Type 2 inhibitor leads of human tropomyosin receptor kinase (hTrkA).
Bioorg.Med.Chem.Lett., 29, 2019
|
|
4Y4S
 
 | | Crystal Structure of Y75A HasA dimer from Yersinia pseudotuberculosis | | Descriptor: | Extracellular heme acquisition hemophore HasA, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Hino, T, Kanadani, M, Muroki, T, Ishimaru, Y, Wada, Y, Sato, T, Ozaki, S. | | Deposit date: | 2015-02-11 | | Release date: | 2015-08-12 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The crystal structure of heme acquisition system A from Yersinia pseudotuberculosis (HasAypt): Roles of the axial ligand Tyr75 and two distal arginines in heme binding
J.Inorg.Biochem., 151, 2015
|
|
6PL3
 
 | | TRK-A IN COMPLEX WITH LIGAND 2a | | Descriptor: | 2-[(3-tert-butyl-1-phenyl-1H-pyrazol-5-yl)amino]-2-oxoethyl 4-(1H-tetrazol-1-yl)benzoate, High affinity nerve growth factor receptor | | Authors: | Subramanian, G. | | Deposit date: | 2019-06-30 | | Release date: | 2019-09-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Type 2 inhibitor leads of human tropomyosin receptor kinase (hTrkA).
Bioorg.Med.Chem.Lett., 29, 2019
|
|
1SBP
 
 | |
1RYW
 
 | | C115S MurA liganded with reaction products | | Descriptor: | GLYCEROL, PHOSPHATE ION, UDP-N-acetylglucosamine 1-carboxyvinyltransferase, ... | | Authors: | Eschenburg, S, Schonbrunn, E. | | Deposit date: | 2003-12-22 | | Release date: | 2004-11-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Evidence That the Fosfomycin Target Cys115 in UDP-N-acetylglucosamine Enolpyruvyl Transferase (MurA) Is Essential for Product Release.
J.Biol.Chem., 280, 2005
|
|
1RX0
 
 | | Crystal structure of isobutyryl-CoA dehydrogenase complexed with substrate/ligand. | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, Acyl-CoA dehydrogenase family member 8, ... | | Authors: | Battaile, K.P, Nguyen, T.V, Vockley, J, Kim, J.J. | | Deposit date: | 2003-12-18 | | Release date: | 2004-04-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structures of Isobutyryl-CoA Dehydrogenase and Enzyme-Product Complex: COMPARISON WITH ISOVALERYL- AND SHORT-CHAIN ACYL-COA DEHYDROGENASES.
J.Biol.Chem., 279, 2004
|
|
