8UVX
 
 | | CosR DNA bound form I | | Descriptor: | DNA (5'-D(*AP*TP*AP*TP*CP*TP*TP*AP*AP*TP*TP*TP*TP*GP*GP*TP*TP*AP*AP*TP*A)-3'), DNA (5'-D(*TP*AP*TP*TP*AP*AP*CP*CP*AP*AP*AP*AP*TP*TP*AP*AP*GP*AP*TP*AP*T)-3'), DNA-binding response regulator | | Authors: | Zhang, Z. | | Deposit date: | 2023-11-05 | | Release date: | 2024-01-31 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis of DNA recognition of the Campylobacter jejuni CosR regulator.
Mbio, 15, 2024
|
|
7U1X
 
 | |
8VPW
 
 | | Pseudomonas fluorescens G150T isocyanide hydratase at 298 K XFEL data, free enzyme | | Descriptor: | CHLORIDE ION, Isonitrile hydratase InhA | | Authors: | Wilson, M.A, Smith, N, Dasgupta, M, Dolamore, C. | | Deposit date: | 2024-01-17 | | Release date: | 2024-02-21 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Changes in an enzyme ensemble during catalysis observed by high-resolution XFEL crystallography.
Sci Adv, 10, 2024
|
|
7LC5
 
 | |
7LAD
 
 | | Clobetasol propionate bound to CYP3A5 | | Descriptor: | Clobetasol propionate, Cytochrome P450 3A5, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Buchman, C.D, Miller, D, Wang, J, Jayaraman, S, Chen, T. | | Deposit date: | 2021-01-06 | | Release date: | 2021-10-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Unraveling the Structural Basis of Selective Inhibition of Human Cytochrome P450 3A5.
J.Am.Chem.Soc., 143, 2021
|
|
7TUS
 
 | |
8F4O
 
 | | Apo structure of the TPP riboswitch aptamer domain | | Descriptor: | IRIDIUM HEXAMMINE ION, TETRAETHYLENE GLYCOL, TPP riboswitch aptamer domain, ... | | Authors: | Lee, H.-K, Wang, Y.-X, Stagno, J.R. | | Deposit date: | 2022-11-11 | | Release date: | 2023-05-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structure of Escherichia coli thiamine pyrophosphate-sensing riboswitch in the apo state.
Structure, 31, 2023
|
|
8VQ1
 
 | | Pseudomonas fluorescens G150T isocyanide hydratase at 298 K XFEL data, thioimidate intermediate | | Descriptor: | CHLORIDE ION, Isonitrile hydratase InhA, N-(4-nitrophenyl)methanimine | | Authors: | Wilson, M.A, Smith, N, Dasgupta, M, Dolamore, C. | | Deposit date: | 2024-01-17 | | Release date: | 2024-02-21 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Changes in an enzyme ensemble during catalysis observed by high-resolution XFEL crystallography.
Sci Adv, 10, 2024
|
|
8ULG
 
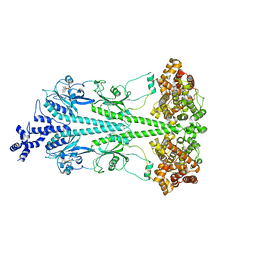 | | Cryo-EM structure of bovine phosphodiesterase 6 bound to IBMX | | Descriptor: | 3-ISOBUTYL-1-METHYLXANTHINE, CYCLIC GUANOSINE MONOPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Aplin, C, Cerione, R.A. | | Deposit date: | 2023-10-16 | | Release date: | 2024-01-17 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Probing the mechanism by which the retinal G protein transducin activates its biological effector PDE6.
J.Biol.Chem., 300, 2023
|
|
8F5T
 
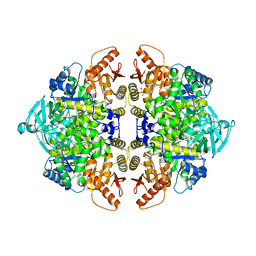 | | Rabbit muscle pyruvate kinase in complex with sodium and magnesium | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Holyoak, T, Fenton, A.W. | | Deposit date: | 2022-11-15 | | Release date: | 2023-05-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | PYK-SubstitutionOME: an integrated database containing allosteric coupling, ligand affinity and mutational, structural, pathological, bioinformatic and computational information about pyruvate kinase isozymes.
Database (Oxford), 2023, 2023
|
|
8F6M
 
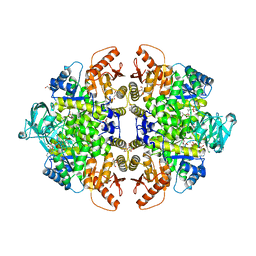 | | Complex of Rabbit muscle pyruvate kinase with ADP and the phosphonate analogue of PEP mimicking the Michaelis complex. | | Descriptor: | (E)-2-METHYL-3-PHOSPHONOACRYLATE, ADENOSINE-5'-DIPHOSPHATE, ALANINE, ... | | Authors: | Holyoak, T, Fenton, A.W. | | Deposit date: | 2022-11-16 | | Release date: | 2023-05-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | PYK-SubstitutionOME: an integrated database containing allosteric coupling, ligand affinity and mutational, structural, pathological, bioinformatic and computational information about pyruvate kinase isozymes.
Database (Oxford), 2023, 2023
|
|
7V9E
 
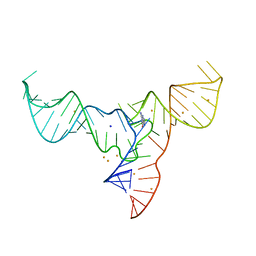 | | Crystal structure of a methyl transferase ribozyme | | Descriptor: | BARIUM ION, GUANINE, RNA (68-MER), ... | | Authors: | Deng, J, Lilley, D.M.J, Huang, L. | | Deposit date: | 2021-08-25 | | Release date: | 2022-03-23 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and mechanism of a methyltransferase ribozyme.
Nat.Chem.Biol., 18, 2022
|
|
7MPZ
 
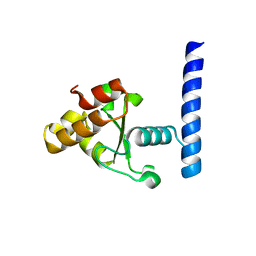 | |
7VFB
 
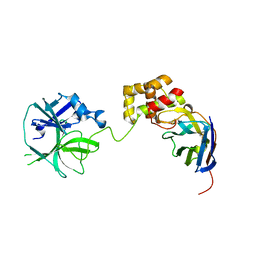 | | the complex of SARS-CoV2 3cl and NB2B4 | | Descriptor: | 3C-like proteinase, nb2b4 | | Authors: | Geng, Y, Sun, Z.C, Wang, L. | | Deposit date: | 2021-09-11 | | Release date: | 2022-03-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | An extended conformation of SARS-CoV-2 main protease reveals allosteric targets.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7V8E
 
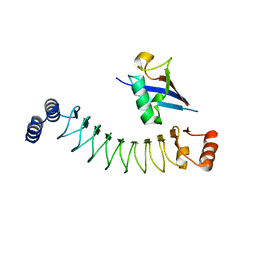 | | Crystal structure of IpaH1.4 LRR domain bound to HOIL-1L UBL domain. | | Descriptor: | RING-type E3 ubiquitin transferase, RanBP-type and C3HC4-type zinc finger-containing protein 1 | | Authors: | Liu, J, Wang, Y, Pan, L. | | Deposit date: | 2021-08-22 | | Release date: | 2022-03-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mechanistic insights into the subversion of the linear ubiquitin chain assembly complex by the E3 ligase IpaH1.4 of Shigella flexneri.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7V8G
 
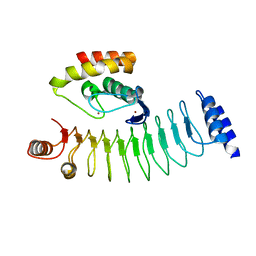 | | Crystal structure of HOIP RING1 domain bound to IpaH1.4 LRR domain | | Descriptor: | E3 ubiquitin-protein ligase RNF31, RING-type E3 ubiquitin transferase, ZINC ION | | Authors: | Liu, J, Wang, Y, Pan, L. | | Deposit date: | 2021-08-23 | | Release date: | 2022-03-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Mechanistic insights into the subversion of the linear ubiquitin chain assembly complex by the E3 ligase IpaH1.4 of Shigella flexneri.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8UGB
 
 | | Cryo-EM structure of bovine phosphodiesterase 6 bound to udenafil | | Descriptor: | CYCLIC GUANOSINE MONOPHOSPHATE, MAGNESIUM ION, Retinal rod rhodopsin-sensitive cGMP 3',5'-cyclic phosphodiesterase subunit gamma, ... | | Authors: | Aplin, C, Cerione, R.A. | | Deposit date: | 2023-10-05 | | Release date: | 2024-01-17 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Probing the mechanism by which the retinal G protein transducin activates its biological effector PDE6.
J.Biol.Chem., 300, 2023
|
|
7V8F
 
 | | Crystal structure of UBE2L3 bound to HOIP RING1 domain. | | Descriptor: | E3 ubiquitin-protein ligase RNF31, Ubiquitin-conjugating enzyme E2 L3, ZINC ION | | Authors: | Liu, J, Wang, Y, Pan, L. | | Deposit date: | 2021-08-22 | | Release date: | 2022-03-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Mechanistic insights into the subversion of the linear ubiquitin chain assembly complex by the E3 ligase IpaH1.4 of Shigella flexneri.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7VFA
 
 | | the complex of SARS-CoV2 3CL and NB1A2 | | Descriptor: | 3C-like proteinase, NB1A2, SULFATE ION | | Authors: | Sun, Z.C, Wang, L, Geng, Y. | | Deposit date: | 2021-09-11 | | Release date: | 2022-03-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | An extended conformation of SARS-CoV-2 main protease reveals allosteric targets.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7XYZ
 
 | | TRIM E3 ubiquitin ligase | | Descriptor: | Tripartite motif-containing protein 72, ZINC ION | | Authors: | Park, S.H, Song, H.K. | | Deposit date: | 2022-06-02 | | Release date: | 2023-07-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (4.62 Å) | | Cite: | Structure and activation of the RING E3 ubiquitin ligase TRIM72 on the membrane.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8U5A
 
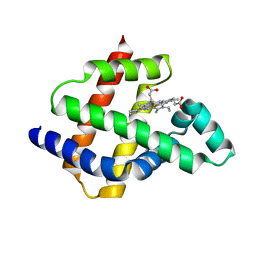 | | Improving protein expression, stability, and function with ProteinMPNN | | Descriptor: | Designed myoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Kalvet, I, Bera, A.K, Baker, D. | | Deposit date: | 2023-09-12 | | Release date: | 2024-01-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Improving Protein Expression, Stability, and Function with ProteinMPNN.
J.Am.Chem.Soc., 146, 2024
|
|
8UGS
 
 | | Cryo-EM structure of bovine phosphodiesterase 6 bound to cGMP | | Descriptor: | CYCLIC GUANOSINE MONOPHOSPHATE, MAGNESIUM ION, Retinal rod rhodopsin-sensitive cGMP 3',5'-cyclic phosphodiesterase subunit gamma, ... | | Authors: | Aplin, C, Cerione, R.A. | | Deposit date: | 2023-10-06 | | Release date: | 2024-01-17 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Probing the mechanism by which the retinal G protein transducin activates its biological effector PDE6.
J.Biol.Chem., 300, 2023
|
|
7VGH
 
 | |
7VED
 
 | |
7V8H
 
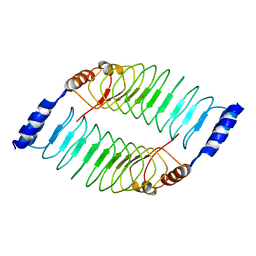 | | Crystal structure of LRR domain from Shigella flexneri IpaH1.4 | | Descriptor: | RING-type E3 ubiquitin transferase | | Authors: | Liu, J, Wang, Y, Pan, L. | | Deposit date: | 2021-08-23 | | Release date: | 2022-03-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Mechanistic insights into the subversion of the linear ubiquitin chain assembly complex by the E3 ligase IpaH1.4 of Shigella flexneri.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
