1UGF
 
 | |
1TXI
 
 | | Crystal structure of the vdr ligand binding domain complexed to TX522 | | Descriptor: | (1R,3R)-5-((Z)-2-((1R,7AS)-HEXAHYDRO-1-((S)-6-HYDROXY-6-METHYLHEPT-4-YN-2-YL)-7A-METHYL-1H-INDEN-4(7AH)-YLIDENE)ETHYLIDENE)CYCLOHEXANE-1,3-DIOL, Vitamin D receptor | | Authors: | Moras, D, Rochel, N. | | Deposit date: | 2004-07-05 | | Release date: | 2005-05-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Superagonistic Action of 14-epi-Analogs of 1,25-Dihydroxyvitamin D Explained by Vitamin D Receptor-Coactivator Interaction
Mol.Pharmacol., 67, 2005
|
|
8B95
 
 | | Crystal structure of PPARG and NCOR2 with BAY-9683, an inverse agonist | | Descriptor: | Nuclear receptor corepressor 2, Peroxisome proliferator-activated receptor gamma, ~{N}1-[[3,4-bis(fluoranyl)phenyl]methyl]-4-chloranyl-6-fluoranyl-~{N}3-(3-methyl-5-morpholin-4-yl-pyridin-2-yl)benzene-1,3-dicarboxamide | | Authors: | Friberg, A, Orsi, D.L, Pook, E, Siegel, S, Lemke, C.T, Stellfeld, T, Puetter, V, Goldstein, J. | | Deposit date: | 2022-10-05 | | Release date: | 2022-12-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Discovery and characterization of orally bioavailable 4-chloro-6-fluoroisophthalamides as covalent PPARG inverse-agonists.
Bioorg.Med.Chem., 78, 2022
|
|
1TXO
 
 | | Crystal structure of the Mycobacterium tuberculosis serine/threonine phosphatase PstP/Ppp at 1.95 A. | | Descriptor: | MANGANESE (II) ION, Putative Bacterial Enzyme | | Authors: | Pullen, K.E, Ng, H.L, Sung, P.Y, Good, M.C, Smith, S.M, Alber, T, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2004-07-05 | | Release date: | 2004-11-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | An Alternate Conformation and a Third Metal in PstP/Ppp, the M. tuberculosis PP2C-Family Ser/Thr Protein Phosphatase.
Structure, 12, 2004
|
|
8B7L
 
 | |
1UBI
 
 | | SYNTHETIC STRUCTURAL AND BIOLOGICAL STUDIES OF THE UBIQUITIN SYSTEM. PART 1 | | Descriptor: | UBIQUITIN | | Authors: | Alexeev, D, Bury, S.M, Turner, M.A, Ogunjobi, O.M, Muir, T.W, Ramage, R, Sawyer, L. | | Deposit date: | 1994-02-03 | | Release date: | 1994-05-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Synthetic, structural and biological studies of the ubiquitin system: the total chemical synthesis of ubiquitin.
Biochem.J., 299, 1994
|
|
8B1U
 
 | | RecBCD-DNA in complex with the phage protein Abc2 and host PpiB | | Descriptor: | Anti-RecBCD protein 2, DNA (70-MER), MAGNESIUM ION, ... | | Authors: | Wilkinson, M, Wilkinson, O.J, Feyerherm, C, Fletcher, E.E, Wigley, D.B, Dillingham, M.S. | | Deposit date: | 2022-09-12 | | Release date: | 2022-12-28 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structures of RecBCD in complex with phage-encoded inhibitor proteins reveal distinctive strategies for evasion of a bacterial immunity hub.
Elife, 11, 2022
|
|
1UBJ
 
 | | Three-dimensional Structure of The Carbon Monoxide Complex of [NiFe]hydrogenase From Desulufovibrio vulgaris Miyazaki F | | Descriptor: | (MU-SULPHIDO)-BIS(MU-CYS,S)-[TRICARBONYLIRON-DI-(CYS,S)NICKEL(II)](FE-NI), CARBON MONOXIDE, FE3-S4 CLUSTER, ... | | Authors: | Ogata, H, Mizoguchi, Y, Mizuno, N, Miki, K, Adachi, S, Yasuoka, N, Yagi, T, Yamauchi, O, Hirota, S, Higuchi, Y. | | Deposit date: | 2003-04-04 | | Release date: | 2003-04-29 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural Studies of the Carbon Monoxide Complex of [NiFe]hydrogenase from Desulfovibrio vulgaris Miyazaki F: Suggestion for the Initial Activation Site for Dihydrogen
J.Am.Chem.Soc., 124, 2002
|
|
8B0C
 
 | | Crystal structure of human heparanase in complex with covalent inhibitor VB158 | | Descriptor: | (1~{S},2~{R},3~{R},4~{S},6~{S})-3,4,6-tris(oxidanyl)-2-[2-[2,2,2-tris(fluoranyl)ethanoylamino]ethoxy]cyclohexane-1-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, Heparanase 50 kDa subunit, ... | | Authors: | Armstrong, Z, Davies, G. | | Deposit date: | 2022-09-07 | | Release date: | 2022-12-28 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | 4-O-Substituted Glucuronic Cyclophellitols are Selective Mechanism-Based Heparanase Inhibitors.
Chemmedchem, 18, 2023
|
|
8B2Q
 
 | | Matrix-metallopeptidase inhibitor Potempin A (PotA) from Tannerella forsythia in complex with T. forsythia karilysin. | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GLYCEROL, ... | | Authors: | Potempa, J, Ksiazek, M, Goulas, T, Cuppari, A, Rodriguez-Banqueri, A, Arolas, J.L, Lopez-Pelegrin, M, Garcia-Ferrer, I, Guevara, T. | | Deposit date: | 2022-09-14 | | Release date: | 2022-12-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | A unique network of attack, defence and competence on the outer membrane of the periodontitis pathogen Tannerella forsythia.
Chem Sci, 14, 2023
|
|
1UCC
 
 | | Crystal structure of the Ribonuclease MC1 from bitter gourd seeds complexed with 3'-UMP. | | Descriptor: | 3'-URIDINEMONOPHOSPHATE, Ribonuclease MC | | Authors: | Suzuki, A, Yao, M, Tanaka, I, Numata, T, Kikukawa, S, Yamasaki, N, Kimura, M. | | Deposit date: | 2003-04-10 | | Release date: | 2003-04-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal structures of the ribonuclease MC1 from bitter gourd seeds, complexed with 2'-UMP or 3'-UMP, reveal structural basis for uridine specificity
Biochem.Biophys.Res.Commun., 275, 2000
|
|
8B0T
 
 | | SARS-CoV-2 Main Protease adduct with Au(PEt3)Br | | Descriptor: | 3C-like proteinase nsp5, GOLD ION | | Authors: | Massai, L, Grifagni, D, De Santis, A, Geri, A, Calderone, V, Cantini, F, Banci, L, Messori, L. | | Deposit date: | 2022-09-08 | | Release date: | 2022-12-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Gold-Based Metal Drugs as Inhibitors of Coronavirus Proteins: The Inhibition of SARS-CoV-2 Main Protease by Auranofin and Its Analogs.
Biomolecules, 12, 2022
|
|
1TY8
 
 | | Crystal structure of yeast ymx7, an ADP-ribose-1''-monophosphatase, complexed with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, Hypothetical 32.1 kDa protein in ADH3-RCA1 intergenic region, ... | | Authors: | Kumaran, D, Swaminathan, S, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2004-07-07 | | Release date: | 2004-11-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | CRYSTAL STRUCTURE OF YEAST HYPOTHETICAL PROTEIN YMX7
To be Published
|
|
1TYH
 
 | |
8B0E
 
 | | Crystal structure of beta-glucuronidase from Acidobacterium capsulatum in complex with covalent inhibitor VB158 | | Descriptor: | (1~{S},2~{R},3~{R},4~{S},6~{S})-3,4,6-tris(oxidanyl)-2-[2-[2,2,2-tris(fluoranyl)ethanoylamino]ethoxy]cyclohexane-1-carboxylic acid, beta-glucuronidase from Acidobacterium capsulatum | | Authors: | Armstrong, Z, Davies, G.J. | | Deposit date: | 2022-09-07 | | Release date: | 2022-12-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | 4-O-Substituted Glucuronic Cyclophellitols are Selective Mechanism-Based Heparanase Inhibitors.
Chemmedchem, 18, 2023
|
|
1TZ2
 
 | | Crystal structure of 1-aminocyclopropane-1-carboyxlate deaminase complexed with ACC | | Descriptor: | 1-AMINOCYCLOPROPANECARBOXYLIC ACID, 1-aminocyclopropane-1-carboxylate deaminase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Karthikeyan, S, Zhou, Q, Zhao, Z, Kao, C.L, Tao, Z, Robinson, H, Liu, H.W, Zhang, H. | | Deposit date: | 2004-07-09 | | Release date: | 2004-11-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Analysis of Pseudomonas 1-Aminocyclopropane-1-carboxylate Deaminase Complexes: Insight into the Mechanism of a Unique Pyridoxal-5'-phosphate Dependent Cyclopropane Ring-Opening Reaction
Biochemistry, 43, 2004
|
|
8B7O
 
 | |
8BLV
 
 | | The PDZ domains of human SDCBP with a bound SDC4 C-terminal peptide | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, Syndecan-4, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Daniel-Mozo, M, Bountra, C, von Delft, F, Brennan, P.E. | | Deposit date: | 2022-11-10 | | Release date: | 2022-12-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The PDZ domains of human SDCBP with a bound SDC4 C-terminal peptide
To Be Published
|
|
1TZS
 
 | | Crystal Structure of an activation intermediate of Cathepsin E | | Descriptor: | 23-mer peptide from PelB-IgG kappa light chain fusion protein, Cathepsin E, activation peptide from Cathepsin E | | Authors: | Ostermann, N, Gerhartz, B, Worpenberg, S, Trappe, J, Eder, J. | | Deposit date: | 2004-07-12 | | Release date: | 2005-07-12 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of an activation intermediate of cathepsin e
J.Mol.Biol., 342, 2004
|
|
1U01
 
 | | High resolution NMR structure of 5-d(GCGT*GCG)-3/5-d(CGCACGC)-3 (T*represents a cyclohexenyl nucleotide) | | Descriptor: | 5'-D(*CP*GP*CP*AP*CP*GP*C)-3', 5'-D(*GP*CP*GP*(XTR)P*GP*CP*G)-3' | | Authors: | Nauwelaerts, K, Lescrinier, E, Sclep, G, Herdewijn, P. | | Deposit date: | 2004-07-12 | | Release date: | 2005-05-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Cyclohexenyl nucleic acids: conformationally flexible oligonucleotides.
Nucleic Acids Res., 33, 2005
|
|
8B8Y
 
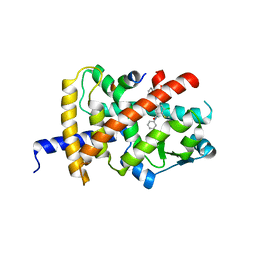 | | Crystal structure of PPARG and NCOR2 with an inverse agonist (compound 7e) | | Descriptor: | 4,5-bis(chloranyl)-~{N}3-phenyl-~{N}1-(phenylmethyl)benzene-1,3-dicarboxamide, Nuclear receptor corepressor 2, Peroxisome proliferator-activated receptor gamma | | Authors: | Holton, S.J, Friberg, A, Orsi, D.L, Pook, E, Siegel, S, Lemke, C.T, Stellfeld, T, Puetter, V, Goldstein, J. | | Deposit date: | 2022-10-05 | | Release date: | 2022-12-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery and characterization of orally bioavailable 4-chloro-6-fluoroisophthalamides as covalent PPARG inverse-agonists.
Bioorg.Med.Chem., 78, 2022
|
|
1U08
 
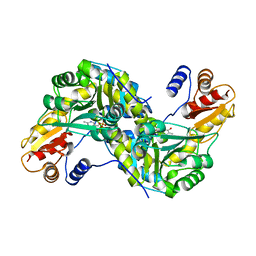 | | Crystal Structure and Reactivity of YbdL from Escherichia coli Identify a Methionine Aminotransferase Function. | | Descriptor: | Hypothetical aminotransferase ybdL, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Dolzan, M, Johansson, K, Roig-Zamboni, V, Campanacci, V, Tegoni, M, Schneider, G, Cambillau, C. | | Deposit date: | 2004-07-13 | | Release date: | 2004-07-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure and reactivity of YbdL from Escherichia coli identify a methionine aminotransferase function
FEBS Lett., 571, 2004
|
|
8BDR
 
 | |
8B93
 
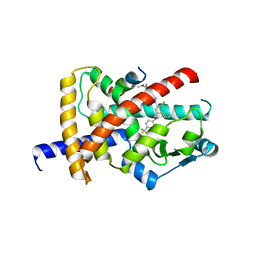 | | Crystal structure of PPARG and NCOR2 with an inverse agonist (compound 15b) | | Descriptor: | 4-chloranyl-6-fluoranyl-~{N}1-[[4-fluoranyl-2-(2-methoxyethoxymethyl)phenyl]methyl]-~{N}3-[2-methyl-4-(trifluoromethyl)phenyl]benzene-1,3-dicarboxamide, Nuclear receptor corepressor 2, Peroxisome proliferator-activated receptor gamma | | Authors: | Friberg, A, Orsi, D.L, Pook, E, Siegel, S, Lemke, C.T, Stellfeld, T, Puetter, V, Goldstein, J. | | Deposit date: | 2022-10-05 | | Release date: | 2022-12-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Discovery and characterization of orally bioavailable 4-chloro-6-fluoroisophthalamides as covalent PPARG inverse-agonists.
Bioorg.Med.Chem., 78, 2022
|
|
1U0M
 
 | | Crystal Structure of 1,3,6,8-Tetrahydroxynaphthalene Synthase (THNS) from Streptomyces coelicolor A3(2): a Bacterial Type III Polyketide Synthase (PKS) Provides Insights into Enzymatic Control of Reactive Polyketide Intermediates | | Descriptor: | GLYCEROL, POLYETHYLENE GLYCOL (N=34), putative polyketide synthase | | Authors: | Austin, M.B, Izumikawa, M, Bowman, M.E, Udwary, D.W, Ferrer, J.L, Moore, B.S, Noel, J.P. | | Deposit date: | 2004-07-13 | | Release date: | 2004-09-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Crystal structure of a bacterial type III polyketide synthase and enzymatic control of reactive polyketide intermediates
J.Biol.Chem., 279, 2004
|
|
