6OCO
 
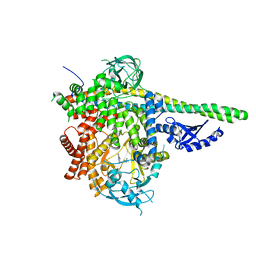 | | HUMAN PI3KDELTA IN COMPLEX WITH COMPOUND 6 | | Descriptor: | 4-[(1S,4S)-5-(3-chlorophenyl)-2,5-diazabicyclo[2.2.1]heptan-2-yl]-2-(pyridin-3-yl)pyrimidine-5-carbonitrile, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform | | Authors: | Lesburg, C.A, Augustin, M.A. | | Deposit date: | 2019-03-25 | | Release date: | 2019-12-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Discovery and optimization of heteroaryl piperazines as potent and selective PI3K delta inhibitors.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
6ZHA
 
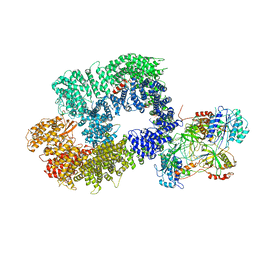 | | Cryo-EM structure of DNA-PK monomer | | Descriptor: | DNA, DNA-dependent protein kinase catalytic subunit,DNA-dependent protein kinase catalytic subunit,DNA-dependent protein kinase catalytic subunit,DNA-PKcs, X-ray repair cross-complementing protein 5, ... | | Authors: | Chaplin, A.K, Hardwick, S.W, Chirgadze, D.Y, Blundell, T.L. | | Deposit date: | 2020-06-21 | | Release date: | 2020-10-21 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.91 Å) | | Cite: | Dimers of DNA-PK create a stage for DNA double-strand break repair.
Nat.Struct.Mol.Biol., 28, 2021
|
|
6PYU
 
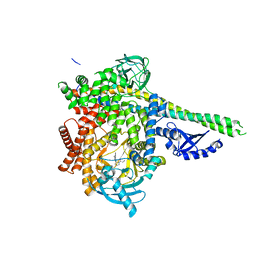 | | Human PI3Kdelta in complex with Compound 4-2 ((3S)-1'-(cyclopropanecarbonyl)-5-(quinoxalin-6-yl)spiro[indole-3,2'-pyrrolidin]-2(1H)-one) | | Descriptor: | (3S)-1'-(cyclopropanecarbonyl)-5-(quinoxalin-6-yl)spiro[indole-3,2'-pyrrolidin]-2(1H)-one, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform | | Authors: | Lesburg, C.A, Augustin, M.A. | | Deposit date: | 2019-07-30 | | Release date: | 2019-08-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Design of selective PI3K delta inhibitors using an iterative scaffold-hopping workflow.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
6Q73
 
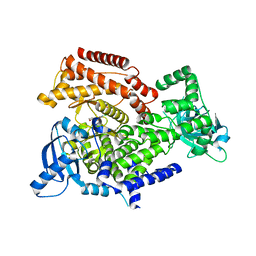 | | PI3K delta in complex with N[2chloro5(3,6dihydro2Hpyran4yl)pyridin3yl]methanesulfonamide | | Descriptor: | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform, ~{N}-[2-chloranyl-5-(3,6-dihydro-2~{H}-pyran-4-yl)pyridin-3-yl]methanesulfonamide | | Authors: | Convery, M.A, Rowland, P, Down, K, Barton, N. | | Deposit date: | 2018-12-12 | | Release date: | 2018-12-26 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Discovery of Potent, Efficient, and Selective Inhibitors of Phosphoinositide 3-Kinase delta through a Deconstruction and Regrowth Approach.
J.Med.Chem., 61, 2018
|
|
7WZW
 
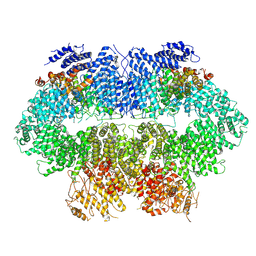 | | Cryo-EM structure of MEC1-DDC2-MMS | | Descriptor: | DNA damage checkpoint protein LCD1, Serine/threonine-protein kinase MEC1 | | Authors: | Zhang, Q, Zhang, Q. | | Deposit date: | 2022-02-19 | | Release date: | 2023-03-01 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structures of Mec1/ATR kinase endogenously stimulated by different genotoxins.
Cell Discov, 8, 2022
|
|
7TYR
 
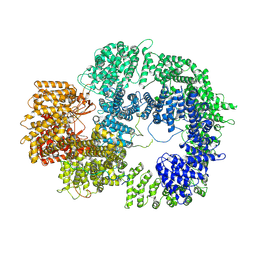 | |
7WZR
 
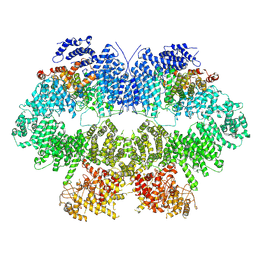 | | Cryo-EM structure of Mec1-HU | | Descriptor: | DNA damage checkpoint protein LCD1, Serine/threonine-protein kinase MEC1 | | Authors: | Zhang, Q, Zhang, Q. | | Deposit date: | 2022-02-19 | | Release date: | 2023-03-08 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Structures of Mec1/ATR kinase endogenously stimulated by different genotoxins.
Cell Discov, 8, 2022
|
|
8TWY
 
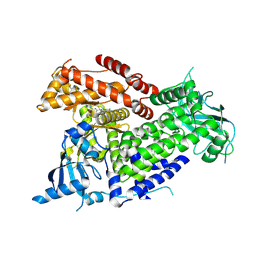 | | Structure of p110 alpha bound to (S)-1-(4-((2-(4-(4-(2-amino-4-(difluoromethyl)pyrimidin-5-yl)-6-(3-methylmorpholino)-1,3,5- triazin-2-yl)piperazin-1-yl)-2-oxoethoxy)methyl)piperidin-1-yl)prop-2-en-1-one (compound 9) | | Descriptor: | 1-(4-{[2-(4-{(4P)-4-[2-amino-4-(difluoromethyl)pyrimidin-5-yl]-6-[(3S)-3-methylmorpholin-4-yl]-1,3,5-triazin-2-yl}piperazin-1-yl)-2-oxoethoxy]methyl}piperidin-1-yl)prop-2-en-1-one, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Burke, J.E, Barlow-Busch, I. | | Deposit date: | 2023-08-21 | | Release date: | 2024-08-28 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Rapid, potent, and persistent covalent chemical probes to deconvolute PI3K alpha signaling.
Chem Sci, 15, 2024
|
|
6YKG
 
 | |
6IG9
 
 | |
6ZFP
 
 | | Cryo-EM structure of DNA-PKcs (State 2) | | Descriptor: | DNA-dependent protein kinase catalytic subunit,DNA-PKcs,DNA-PKcs | | Authors: | Chaplin, A.K, Hardwick, S.W, Chirgadze, D.Y, Blundell, T.L. | | Deposit date: | 2020-06-17 | | Release date: | 2020-10-21 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | Dimers of DNA-PK create a stage for DNA double-strand break repair.
Nat.Struct.Mol.Biol., 28, 2021
|
|
6ZAA
 
 | | PI3K Delta in complex with methoxy(methylsulfamoyl)pyridinylN(methylpiperidinyl)dihydrobenzoxazinecarboxamide | | Descriptor: | 4-[6-methoxy-5-(methylsulfamoyl)pyridin-3-yl]-~{N}-(1-methylpiperidin-4-yl)-2,3-dihydro-1,4-benzoxazine-6-carboxamide, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform | | Authors: | Convery, M.A, Hardy, C.J, Spencer, J.A, Rowland, P. | | Deposit date: | 2020-06-05 | | Release date: | 2020-07-15 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Design and Development of a Macrocyclic Series Targeting Phosphoinositide 3-Kinase delta.
Acs Med.Chem.Lett., 11, 2020
|
|
6ZHE
 
 | | Cryo-EM structure of DNA-PK dimer | | Descriptor: | DNA (25-MER), DNA (26-MER), DNA (27-MER), ... | | Authors: | Chaplin, A.K, Hardwick, S.W, Chirgadze, D.Y, Blundell, T.L. | | Deposit date: | 2020-06-23 | | Release date: | 2020-10-21 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (7.24 Å) | | Cite: | Dimers of DNA-PK create a stage for DNA double-strand break repair.
Nat.Struct.Mol.Biol., 28, 2021
|
|
5O83
 
 | | Discovery of CDZ173 (leniolisib), Representing a Structurally Novel Class of PI3K Delta-Selective Inhibitors | | Descriptor: | Leniolisib, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform | | Authors: | Gutmann, S, Rummel, G, Shrestha, B. | | Deposit date: | 2017-06-12 | | Release date: | 2017-09-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Discovery of CDZ173 (Leniolisib), Representing a Structurally Novel Class of PI3K Delta-Selective Inhibitors.
ACS Med Chem Lett, 8, 2017
|
|
6ZH2
 
 | | Cryo-EM structure of DNA-PKcs (State 1) | | Descriptor: | DNA-dependent protein kinase catalytic subunit,DNA-dependent protein kinase catalytic subunit,DNA-dependent protein kinase catalytic subunit,DNA-dependent protein kinase catalytic subunit,DNA-dependent protein kinase catalytic subunit,DNA-dependent protein kinase catalytic subunit,DNA-dependent protein kinase catalytic subunit,DNA-dependent protein kinase catalytic subunit,DNA-PKcs | | Authors: | Chaplin, A.K, Hardwick, S.W, Chirgadze, D.Y, Blundell, T.L. | | Deposit date: | 2020-06-20 | | Release date: | 2020-10-21 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.92 Å) | | Cite: | Dimers of DNA-PK create a stage for DNA double-strand break repair.
Nat.Struct.Mol.Biol., 28, 2021
|
|
4XE0
 
 | | Idelalisib bound to the p110 subunit of PI3K delta | | Descriptor: | 5-fluoro-3-phenyl-2-[(1S)-1-(7H-purin-6-ylamino)propyl]quinazolin-4(3H)-one, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform | | Authors: | Somoza, J.R, Villasenor, A. | | Deposit date: | 2014-12-20 | | Release date: | 2015-02-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.434 Å) | | Cite: | Structural, Biochemical, and Biophysical Characterization of Idelalisib Binding to Phosphoinositide 3-Kinase delta.
J.Biol.Chem., 290, 2015
|
|
6ZAC
 
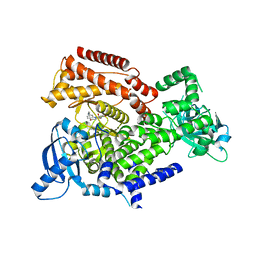 | | PI3K Delta in complex with [(dimethylamino)methyldihydrobenzoxazin2methoxypyridinyl]methanesulfonamide | | Descriptor: | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform, ~{N}-[5-[6-[(dimethylamino)methyl]-2,3-dihydro-1,4-benzoxazin-4-yl]-2-methoxy-pyridin-3-yl]methanesulfonamide | | Authors: | Convery, M.A, Hardy, C.J, Spencer, J.A, Rowland, P. | | Deposit date: | 2020-06-05 | | Release date: | 2020-07-01 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Design and Development of a Macrocyclic Series Targeting Phosphoinositide 3-Kinase delta.
Acs Med.Chem.Lett., 11, 2020
|
|
6ZH4
 
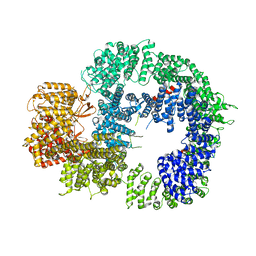 | | Cryo-EM structure of DNA-PKcs (State 3) | | Descriptor: | DNA-dependent protein kinase catalytic subunit,DNA-PKcs | | Authors: | Chaplin, A.K, Hardwick, S.W, Chirgadze, D.Y, Blundell, T.L. | | Deposit date: | 2020-06-20 | | Release date: | 2020-10-21 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.62 Å) | | Cite: | Dimers of DNA-PK create a stage for DNA double-strand break repair.
Nat.Struct.Mol.Biol., 28, 2021
|
|
6ZH6
 
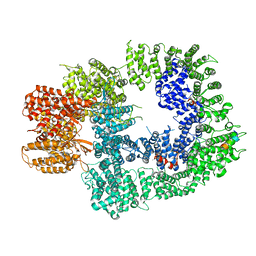 | | Cryo-EM structure of DNA-PKcs:Ku80ct194 | | Descriptor: | DNA-dependent protein kinase catalytic subunit,DNA-PKcs, X-ray repair cross-complementing protein 5 | | Authors: | Chaplin, A.K, Hardwick, S.W, Chirgadze, D.Y, Blundell, T.L. | | Deposit date: | 2020-06-21 | | Release date: | 2020-10-21 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.93 Å) | | Cite: | Dimers of DNA-PK create a stage for DNA double-strand break repair.
Nat.Struct.Mol.Biol., 28, 2021
|
|
6Z3A
 
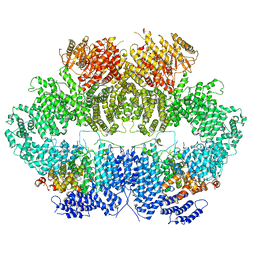 | | Mec1-Ddc2 (wild-type) in complex with AMP-PNP | | Descriptor: | DNA damage checkpoint protein LCD1, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Serine/threonine-protein kinase MEC1, ... | | Authors: | Yates, L.A, Zhang, X. | | Deposit date: | 2020-05-19 | | Release date: | 2020-11-11 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Mechanism of auto-inhibition and activation of Mec1 ATR checkpoint kinase.
Nat.Struct.Mol.Biol., 28, 2021
|
|
4XX5
 
 | | Structure of PI3K gamma in complex with an inhibitor | | Descriptor: | N-[5-(5-methoxypyridin-3-yl)-4,5,6,7-tetrahydro[1,3]thiazolo[5,4-c]pyridin-2-yl]acetamide, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform | | Authors: | Collier, P.N, Messersmith, D, Le Tiran, A, Bandarage, U.K, Boucher, C, Come, J, Cottrell, K.M, Damagnez, V, Doran, J.D, Griffith, J.P, Khare-Pandit, S, Krueger, E.B, Ledeboer, M.W, Ledford, B, Liao, Y, Mahajan, S, Moody, C.S, Wang, T, Xu, J, Aronov, A.M. | | Deposit date: | 2015-01-29 | | Release date: | 2015-08-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Structure of PI3K gamma in complex with an inhibitor
To Be Published
|
|
5NCY
 
 | | mPI3Kd IN COMPLEX WITH inh1 | | Descriptor: | 1,2-ETHANEDIOL, 4-azanyl-6-[[(1~{S})-1-(3-methyl-5-oxidanylidene-6-phenyl-[1,3]thiazolo[3,2-a]pyridin-7-yl)ethyl]amino]pyrimidine-5-carbonitrile, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform, ... | | Authors: | Petersen, J. | | Deposit date: | 2017-03-06 | | Release date: | 2017-06-21 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Design and Synthesis of Soluble and Cell-Permeable PI3K delta Inhibitors for Long-Acting Inhaled Administration.
J. Med. Chem., 60, 2017
|
|
4XZ4
 
 | | Structure of PI3K gamma in complex with an inhibitor | | Descriptor: | N-[5-(6-methoxypyrazin-2-yl)-4,5,6,7-tetrahydro[1,3]thiazolo[5,4-c]pyridin-2-yl]acetamide, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform | | Authors: | Collier, P.N, Messersmith, D, Le Tiran, A, Bandarage, U.K, Boucher, C, Come, J, Cottrell, K.M, Damagnez, V, Doran, J.D, Griffith, J.P, Khare-Pandit, S, Krueger, E.B, Ledeboer, M.W, Ledford, B, Liao, Y, Mahajan, S, Moody, C.S, Wang, T, Xu, J, Aronov, A.M. | | Deposit date: | 2015-02-03 | | Release date: | 2016-02-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of PI3K gamma in complex with an inhibitor
To Be Published
|
|
5I0N
 
 | | PI4K IIalpha bound to calcium | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, Phosphatidylinositol 4-kinase type 2-alpha,Lysozyme,Phosphatidylinositol 4-kinase type 2-alpha | | Authors: | Baumlova, A, Boura, E. | | Deposit date: | 2016-02-04 | | Release date: | 2016-03-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | PI4K IIalpha bound to calcium
To Be Published
|
|
5OJS
 
 | |
