3KQB
 
 | | Factor xa in complex with the inhibitor n-(3-fluoro-2'- (methylsulfonyl)biphenyl-4-yl)-1-(3-(5-oxo-4,5-dihydro-1h- 1,2,4-triazol-3-yl)phenyl)-3-(trifluoromethyl)-1h- pyrazole-5-carboxamide | | Descriptor: | N-(3-FLUORO-2'-(METHYLSULFONYL)BIPHENYL-4-YL)-1-(3-(5-OXO-4,5-DIHYDRO-1H-1,2,4-TRIAZOL-3-YL)PHENYL)-3-(TRIFLUOROMETHYL)-1H-PYRAZOLE-5-CARBOXAMIDE, SODIUM ION, factor Xa heavy chain, ... | | Authors: | Sheriff, S. | | Deposit date: | 2009-11-17 | | Release date: | 2010-02-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Phenyltriazolinones as potent factor Xa inhibitors.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3KQL
 
 | |
3KVR
 
 | | Trapping of an oxocarbenium ion intermediate in UP crystals | | Descriptor: | 2,5-anhydro-4-deoxy-D-erythro-pent-4-enitol, 5-FLUOROURACIL, SULFATE ION, ... | | Authors: | Paul, D, O'Leary, S, Rajashankar, K, Bu, W, Toms, A, Settembre, E, Sanders, J, Begley, T.P, Ealick, S.E. | | Deposit date: | 2009-11-30 | | Release date: | 2010-04-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Glycal formation in crystals of uridine phosphorylase.
Biochemistry, 49, 2010
|
|
3KQY
 
 | |
3KW3
 
 | |
3KSX
 
 | |
3KYA
 
 | |
3KT8
 
 | | Crystal structure of S. cerevisiae tryptophanyl-tRNA synthetase in complex with L-tryptophanamide | | Descriptor: | L-TRYPTOPHANAMIDE, SULFATE ION, Tryptophanyl-tRNA synthetase, ... | | Authors: | Zhou, M, Dong, X, Zhong, C, Shen, N, Ding, J. | | Deposit date: | 2009-11-24 | | Release date: | 2010-02-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structures of Saccharomyces cerevisiae tryptophanyl-tRNA synthetase: new insights into the mechanism of tryptophan activation and implications for anti-fungal drug design
Nucleic Acids Res., 38, 2010
|
|
3KU1
 
 | |
3KYY
 
 | |
3KW4
 
 | | Crystal structure of cytochrome 2B4 in complex with the anti-platelet drug ticlopidine | | Descriptor: | 2-{[(3alpha,5alpha,7alpha,8alpha,10alpha,12alpha,17alpha)-3,12-bis{2-[(4-O-alpha-D-glucopyranosyl-beta-D-glucopyranosyl)oxy]ethoxy}cholan-7-yl]oxy}ethyl 4-O-alpha-D-glucopyranosyl-beta-D-glucopyranoside, 5-CYCLOHEXYL-1-PENTYL-BETA-D-MALTOSIDE, Cytochrome P450 2B4, ... | | Authors: | Gay, S.C, Maekawa, K, Roberts, A.G, Hong, W.-X, Zhang, Q, Stout, C.D, Halpert, J.R. | | Deposit date: | 2009-11-30 | | Release date: | 2010-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Structures of cytochrome P450 2B4 complexed with the antiplatelet drugs ticlopidine and clopidogrel.
Biochemistry, 49, 2010
|
|
3KWK
 
 | |
3KXM
 
 | | Crystal structure of Z. mays CK2 kinase alpha subunit in complex with the inhibitor K74 | | Descriptor: | Casein kinase II subunit alpha, N-methyl-2-[(4,5,6,7-tetrabromo-1-methyl-1H-benzimidazol-2-yl)sulfanyl]acetamide | | Authors: | Papinutto, E, Franchin, C, Battistutta, R. | | Deposit date: | 2009-12-03 | | Release date: | 2010-11-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | ATP site-directed inhibitors of protein kinase CK2: an update.
Curr Top Med Chem, 11, 2011
|
|
3L1F
 
 | | Bovine AlphaA crystallin | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Alpha-crystallin A chain | | Authors: | Laganowsky, A, Sawaya, M.R, Cascio, D, Eisenberg, D. | | Deposit date: | 2009-12-11 | | Release date: | 2010-05-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Crystal structures of truncated alphaA and alphaB crystallins reveal structural mechanisms of polydispersity important for eye lens function.
Protein Sci., 19, 2010
|
|
3KYK
 
 | | Crystal structure of li33 Igg1 Fab | | Descriptor: | Heavy Chain Li33 IgG1, Light Chain Li33 IgG1, SULFATE ION | | Authors: | Silvian, L.F, Pepinsky, R.B, Walus, L. | | Deposit date: | 2009-12-06 | | Release date: | 2010-03-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Improving the solubility of anti-LINGO-1 monoclonal antibody Li33 by isotype switching and targeted mutagenesis.
Protein Sci., 19, 2010
|
|
3L20
 
 | | Crystal structure of a hypothetical protein from Staphylococcus aureus | | Descriptor: | Putative uncharacterized protein | | Authors: | Lam, R, Chan, T, Battaile, K.P, Mihajlovic, V, Romanov, V, Soloveychik, M, Kisselman, G, McGrath, T.E, Lam, K, Pai, E.F, Chirgadze, N.Y. | | Deposit date: | 2009-12-14 | | Release date: | 2010-10-27 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.451 Å) | | Cite: | Crystal structure of a hypothetical protein from Staphylococcus aureus
To be Published
|
|
3NHP
 
 | |
3NIH
 
 | | The structure of UBR box (RIAAA) | | Descriptor: | E3 ubiquitin-protein ligase UBR1, Peptide RIAAA, ZINC ION | | Authors: | Choi, W.S, Jeong, B.-C, Lee, M.-R, Song, H.K. | | Deposit date: | 2010-06-16 | | Release date: | 2010-09-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for the recognition of N-end rule substrates by the UBR box of ubiquitin ligases
Nat.Struct.Mol.Biol., 17, 2010
|
|
3NJH
 
 | | D37A mutant of SO1698 protein, an aspartic peptidase from Shewanella oneidensis. | | Descriptor: | CALCIUM ION, GLYCEROL, Peptidase, ... | | Authors: | Osipiuk, J, Mulligan, R, Bargassa, M, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-06-17 | | Release date: | 2010-07-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Characterization of member of DUF1888 protein family, self-cleaving and self-assembling endopeptidase.
J.Biol.Chem., 287, 2012
|
|
3NJP
 
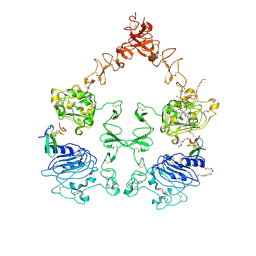 | | The Extracellular and Transmembrane Domain Interfaces in Epidermal Growth Factor Receptor Signaling | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Epidermal growth factor, Epidermal growth factor receptor, ... | | Authors: | Lu, C, Mi, L.-Z, Grey, M.J, Zhu, J, Graef, E, Yokoyama, S, Springer, T.A. | | Deposit date: | 2010-06-17 | | Release date: | 2010-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.304 Å) | | Cite: | Structural evidence for loose linkage between ligand binding and kinase activation in the epidermal growth factor receptor.
Mol.Cell.Biol., 30, 2010
|
|
3NM1
 
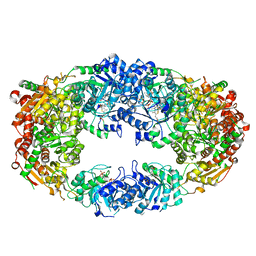 | | The Crystal Structure of Candida glabrata THI6, a Bifunctional Enzyme involved in Thiamin Biosyhthesis of Eukaryotes | | Descriptor: | 2-TRIFLUOROMETHYL-5-METHYLENE-5H-PYRIMIDIN-4-YLIDENEAMINE, 4-methyl-5-[2-(phosphonooxy)ethyl]-1,3-thiazole-2-carboxylic acid, MAGNESIUM ION, ... | | Authors: | Paul, D, Chatterjee, A, Begley, T.P, Ealick, S.E. | | Deposit date: | 2010-06-21 | | Release date: | 2010-11-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.211 Å) | | Cite: | Domain Organization in Candida glabrata THI6, a Bifunctional Enzyme Required for Thiamin Biosynthesis in Eukaryotes .
Biochemistry, 49, 2010
|
|
3MYQ
 
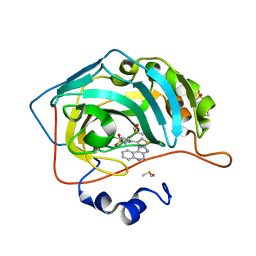 | | Crystal structure of human carbonic anhydrase isozyme II with 2-chloro-5-[(1H-imidazo[4,5-c]quinolin-2-ylsulfanyl)acetyl]benzenesulfonamide | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-chloro-5-[(1H-imidazo[4,5-c]quinolin-2-ylsulfanyl)acetyl]benzenesulfonamide, Carbonic anhydrase 2, ... | | Authors: | Grazulis, S, Manakova, E, Golovenko, D. | | Deposit date: | 2010-05-11 | | Release date: | 2011-02-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Indapamide-like benzenesulfonamides as inhibitors of carbonic anhydrases I, II, VII, and XIII.
Bioorg.Med.Chem., 18, 2010
|
|
3N6X
 
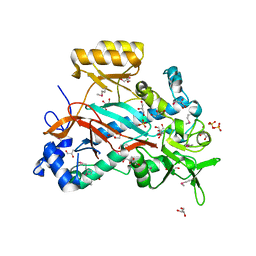 | |
3NKB
 
 | | A 1.9A crystal structure of the HDV ribozyme precleavage suggests both Lewis acid and general acid mechanisms contribute to phosphodiester cleavage | | Descriptor: | DNA/RNA (5'-D(*(DUR))-D(*GP*G)-R(P*CP*UP*UP*GP*CP*A)-3'), MAGNESIUM ION, The hepatitis delta virus ribozyme | | Authors: | Chen, J.-H, Yajima, R, Chadalavada, D.M, Chase, E, Bevilacqua, P.C, Golden, B.L. | | Deposit date: | 2010-06-18 | | Release date: | 2010-09-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.916 Å) | | Cite: | A 1.9 A crystal structure of the HDV ribozyme precleavage suggests both Lewis acid and general acid mechanisms contribute to phosphodiester cleavage.
Biochemistry, 49, 2010
|
|
3MWX
 
 | |
