6K9T
 
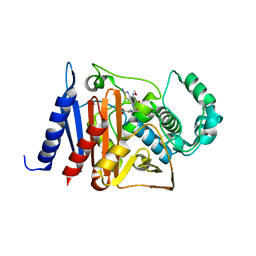 | | Crystal structure of a class C beta-lactamase in complex with cefotaxime | | Descriptor: | Beta-lactamase, CEFOTAXIME, C3' cleaved, ... | | Authors: | Bae, D.W, Jung, Y.E, An, Y.J, Na, J.H, Cha, S.S. | | Deposit date: | 2019-06-17 | | Release date: | 2019-10-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.459 Å) | | Cite: | Structural Insights into Catalytic Relevances of Substrate Poses in ACC-1.
Antimicrob.Agents Chemother., 63, 2019
|
|
2G1D
 
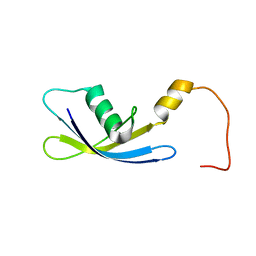 | | Solution Structure of Ribosomal Protein S24E from Thermoplasma acidophilum | | Descriptor: | 30S ribosomal protein S24e | | Authors: | Jeon, B.-Y, Hong, E.-M, Jung, J.-W, Yee, A, Arrowsmith, C.H, Lee, W. | | Deposit date: | 2006-02-14 | | Release date: | 2007-02-14 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of TA1092, a ribosomal protein S24e from Thermoplasma acidophilum
Proteins, 64, 2006
|
|
2G67
 
 | |
3R5L
 
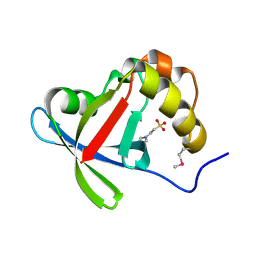 | | Structure of Ddn, the Deazaflavin-dependent nitroreductase from Mycobacterium tuberculosis involved in bioreductive activation of PA-824 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Deazaflavin-dependent nitroreductase | | Authors: | Cellitti, S.E, Shaffer, J, Jones, D.H, Mukherjee, T, Gurumurthy, M, Bursulaya, B, Boshoff, H.I.M, Choi, I, Nayyar, A, Lee, Y.S, Cherian, J, Niyomrattanakit, P, Dick, T, Manjunatha, U.H, Barry, C.E, Spraggon, G, Geierstanger, B.H. | | Deposit date: | 2011-03-18 | | Release date: | 2012-01-18 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure of Ddn, the deazaflavin-dependent nitroreductase from Mycobacterium tuberculosis involved in bioreductive activation of PA-824.
Structure, 20, 2012
|
|
3R5R
 
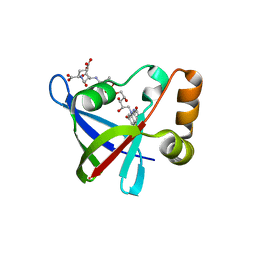 | | Structure of Ddn, the Deazaflavin-dependent nitroreductase from Mycobacterium tuberculosis involved in bioreductive activation of PA-824, with co-factor F420 | | Descriptor: | COENZYME F420, Deazaflavin-dependent nitroreductase | | Authors: | Cellitti, S.E, Shaffer, J, Jones, D.H, Mukherjee, T, Gurumurthy, M, Bursulaya, B, Boshoff, H.I.M, Choi, I, Nayya, A, Lee, Y.S, Cherian, J, Niyomrattanakit, P, Dick, T, Manjunatha, U.H, Barry, C.E, Spraggon, G, Geierstanger, B.H. | | Deposit date: | 2011-03-19 | | Release date: | 2012-01-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Structure of Ddn, the deazaflavin-dependent nitroreductase from Mycobacterium tuberculosis involved in bioreductive activation of PA-824.
Structure, 20, 2012
|
|
3R5W
 
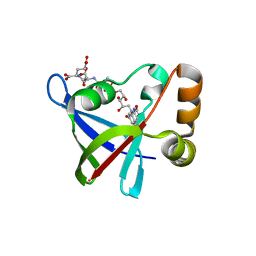 | | Structure of Ddn, the Deazaflavin-dependent nitroreductase from Mycobacterium tuberculosis involved in bioreductive activation of PA-824, with co-factor F420 | | Descriptor: | COENZYME F420, Deazaflavin-dependent nitroreductase | | Authors: | Cellitti, S.E, Shaffer, J, Jones, D.H, Mukherjee, T, Gurumurthy, M, Bursulaya, B, Boshoff, H.I.M, Choi, I, Nayya, A, Lee, Y.S, Cherian, J, Niyomrattanakit, P, Dick, T, Manjunatha, U.H, Barry, C.E, Spraggon, G, Geierstanger, B.H. | | Deposit date: | 2011-03-20 | | Release date: | 2012-01-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.786 Å) | | Cite: | Structure of Ddn, the deazaflavin-dependent nitroreductase from Mycobacterium tuberculosis involved in bioreductive activation of PA-824.
Structure, 20, 2012
|
|
3R5P
 
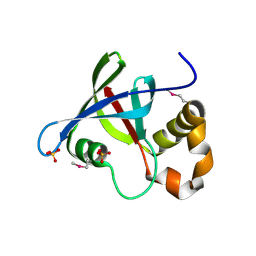 | | Structure of Ddn, the Deazaflavin-dependent nitroreductase from Mycobacterium tuberculosis involved in bioreductive activation of PA-824 | | Descriptor: | Deazaflavin-dependent nitroreductase, SULFATE ION | | Authors: | Cellitti, S.E, Shaffer, J, Jones, D.H, Mukherjee, T, Gurumurthy, M, Bursulaya, B, Boshoff, H.I.M, Choi, I, Nayya, A, Lee, Y.S, Cherian, J, Niyomrattanakit, P, Dick, T, Manjunatha, U.H, Barry, C.E, Spraggon, G, Geierstanger, B.H. | | Deposit date: | 2011-03-19 | | Release date: | 2012-01-18 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of Ddn, the deazaflavin-dependent nitroreductase from Mycobacterium tuberculosis involved in bioreductive activation of PA-824.
Structure, 20, 2012
|
|
3R5Y
 
 | | Structure of a Deazaflavin-dependent nitroreductase from Nocardia farcinica, with co-factor F420 | | Descriptor: | COENZYME F420, Putative uncharacterized protein | | Authors: | Cellitti, S.E, Shaffer, J, Jones, D.H, Mukherjee, T, Gurumurthy, M, Bursulaya, B, Boshoff, H.I.M, Choi, I, Nayya, A, Lee, Y.S, Cherian, J, Niyomrattanakit, P, Dick, T, Manjunatha, U.H, Barry, C.E, Spraggon, G, Geierstanger, B.H. | | Deposit date: | 2011-03-20 | | Release date: | 2012-01-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Structure of Ddn, the deazaflavin-dependent nitroreductase from Mycobacterium tuberculosis involved in bioreductive activation of PA-824.
Structure, 20, 2012
|
|
6FJS
 
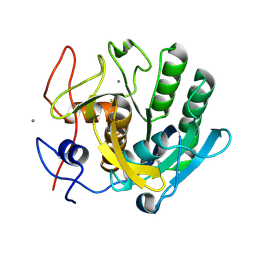 | | Proteinase~K SIRAS phased structure of room-temperature, serially collected synchrotron data | | Descriptor: | CALCIUM ION, Proteinase K | | Authors: | Botha, S, Baitan, D, Jungnickel, K.E.J, Oberthuer, D, Schmidt, C, Stern, S, Wiedorn, M.O, Perbandt, M, Chapman, H.N, Betzel, C. | | Deposit date: | 2018-01-23 | | Release date: | 2018-10-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | De novoprotein structure determination by heavy-atom soaking in lipidic cubic phase and SIRAS phasing using serial synchrotron crystallography.
IUCrJ, 5, 2018
|
|
3Q8J
 
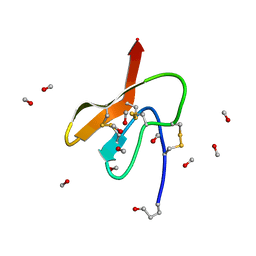 | | Crystal Structure of Asteropsin A from Marine Sponge Asteropus sp. | | Descriptor: | Asteropsin A, METHANOL | | Authors: | Bowling, J.J, Fronczek, F.R, Hamann, M.T, Li, H, Jung, J.H. | | Deposit date: | 2011-01-06 | | Release date: | 2012-04-18 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (0.87 Å) | | Cite: | An Uncommon Crystal Structure of a Marine Knottin Peptide from Asteropus sp.
To be published
|
|
3R5Z
 
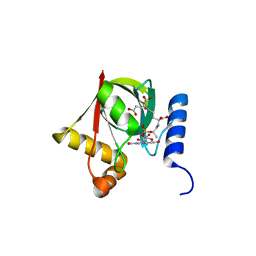 | | Structure of a Deazaflavin-dependent reductase from Nocardia farcinica, with co-factor F420 | | Descriptor: | COENZYME F420, Putative uncharacterized protein, SULFATE ION | | Authors: | Cellitti, S.E, Shaffer, J, Jones, D.H, Mukherjee, T, Gurumurthy, M, Bursulaya, B, Boshoff, H.I.M, Choi, I, Nayya, A, Lee, Y.S, Cherian, J, Niyomrattanakit, P, Dick, T, Manjunatha, U.H, Barry, C.E, Spraggon, G, Geierstanger, B.H. | | Deposit date: | 2011-03-20 | | Release date: | 2012-01-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.503 Å) | | Cite: | Structure of Ddn, the deazaflavin-dependent nitroreductase from Mycobacterium tuberculosis involved in bioreductive activation of PA-824.
Structure, 20, 2012
|
|
6ED7
 
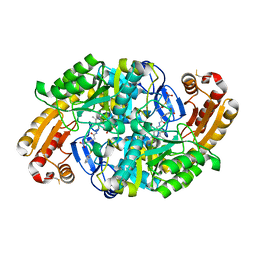 | | Crystal structure of 7,8-diaminopelargonic acid synthase bound to inhibitor MAC13772 | | Descriptor: | 2-[(2-nitrophenyl)sulfanyl]acetohydrazide, 7,8-diamino-pelargonic acid aminotransferase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Brown, C.M, Zlitni, S, Chan, J, Brown, E.D, Junop, M.S. | | Deposit date: | 2018-08-08 | | Release date: | 2019-08-21 | | Last modified: | 2020-01-08 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Crystal structure of 7,8-diaminopelargonic acid synthase bound to inhibitor MAC13772
To Be Published
|
|
4PZO
 
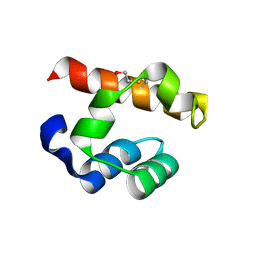 | | Crystal structure of PHC3 SAM L967R | | Descriptor: | Polyhomeotic-like protein 3 | | Authors: | Nanyes, D.R, Junco, S.E, Taylor, A.B, Robinson, A.K, Patterson, N.L, Shivarajpur, A, Halloran, J, Hale, S.M, Kaur, Y, Hart, P.J, Kim, C.A. | | Deposit date: | 2014-03-31 | | Release date: | 2014-07-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Multiple polymer architectures of human polyhomeotic homolog 3 sterile alpha motif.
Proteins, 82, 2014
|
|
8RIJ
 
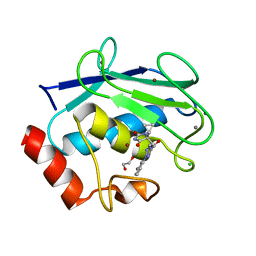 | | Discovery of the first orally bioavailable ADAMTS7 inhibitor BAY-9835 | | Descriptor: | CALCIUM ION, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Schafer, M, Meibom, D, Wasnaire, P, Beyer, K, Broehl, A, Cancho-Grande, Y, Elowe, N, Henninger, K, Johannes, S, Jungmann, N, Krainz, T, Lindner, N, Maassen, S, MacDonald, B, Menshykau, D, Mittendorf, J, Sanchez, G, Stefan, E, Torge, A, Xing, Y, Zubov, D. | | Deposit date: | 2023-12-18 | | Release date: | 2024-02-21 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | BAY-9835: Discovery of the First Orally Bioavailable ADAMTS7 Inhibitor.
J.Med.Chem., 67, 2024
|
|
4PZN
 
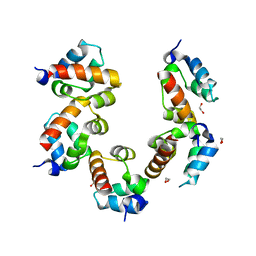 | | Crystal structure of PHC3 SAM L971E | | Descriptor: | 1,2-ETHANEDIOL, Polyhomeotic-like protein 3 | | Authors: | Nanyes, D.R, Junco, S.E, Taylor, A.B, Robinson, A.K, Patterson, N.L, Shivarajpur, A, Halloran, J, Hale, S.M, Kaur, Y, Hart, P.J, Kim, C.A. | | Deposit date: | 2014-03-31 | | Release date: | 2014-07-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Multiple polymer architectures of human polyhomeotic homolog 3 sterile alpha motif.
Proteins, 82, 2014
|
|
1FF2
 
 | |
8R1A
 
 | | Model of the membrane-bound GBP1 oligomer | | Descriptor: | ALUMINUM FLUORIDE, GUANOSINE-5'-DIPHOSPHATE, Guanylate binding protein 1, ... | | Authors: | Weismehl, M, Chu, X, Kutsch, M, Lauterjung, P, Herrmann, C, Kudryashev, M, Daumke, O. | | Deposit date: | 2023-11-01 | | Release date: | 2024-01-17 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (26.799999 Å) | | Cite: | Structural insights into the activation mechanism of antimicrobial GBP1.
Embo J., 43, 2024
|
|
4RMG
 
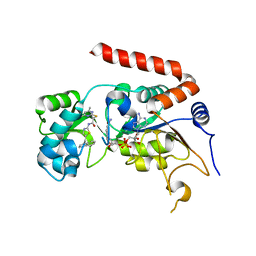 | | Human Sirt2 in complex with SirReal2 and NAD+ | | Descriptor: | 2-[(4,6-dimethylpyrimidin-2-yl)sulfanyl]-N-[5-(naphthalen-1-ylmethyl)-1,3-thiazol-2-yl]acetamide, NAD-dependent protein deacetylase sirtuin-2, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Rumpf, T, Schiedel, M, Karaman, B, Roessler, C, North, B.J, Lehotzky, A, Olah, J, Ladwein, K.I, Schmidtkunz, K, Gajer, M, Pannek, M, Steegborn, C, Sinclair, D.A, Gerhardt, S, Ovadi, J, Schutkowski, M, Sippl, W, Einsle, O, Jung, M. | | Deposit date: | 2014-10-21 | | Release date: | 2015-02-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Selective Sirt2 inhibition by ligand-induced rearrangement of the active site.
Nat Commun, 6, 2015
|
|
8SQ7
 
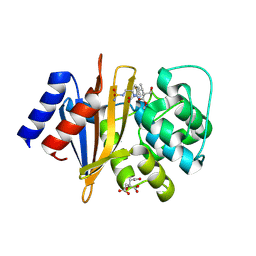 | | X-ray crystal structure of Acinetobacter baumanii beta-lactamase variant OXA-82 K83D in complex with doripenem | | Descriptor: | (4R,5S)-5-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-4-methyl-3-({(3S,5S)-5-[(sulfamoylamino)methyl]pyrrolidin-3-yl}sulfanyl)-4,5-dihydro-1H-pyrrole-2-carboxylic acid, Beta-lactamase OXA-82, CITRATE ANION, ... | | Authors: | Powers, R.A, Leonard, D.A, June, C.M, Szarecka, A, Wawrzak, Z. | | Deposit date: | 2023-05-04 | | Release date: | 2024-05-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structural and Dynamic Features of Acinetobacter baumannii OXA-66 beta-Lactamase Explain Its Stability and Evolution of Novel Variants.
J.Mol.Biol., 436, 2024
|
|
8SQ8
 
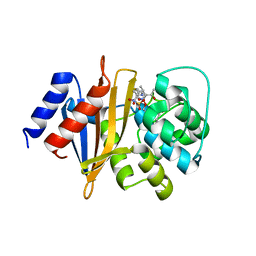 | | X-ray crystal structure of Acinetobacter baumanii beta-lactamase variant OXA-109 in complex with doripenem | | Descriptor: | (4R,5S)-5-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-4-methyl-3-({(3S,5S)-5-[(sulfamoylamino)methyl]pyrrolidin-3-yl}sulfanyl)-4,5-dihydro-1H-pyrrole-2-carboxylic acid, BICARBONATE ION, Beta-lactamase OXA-109 | | Authors: | Powers, R.A, Leonard, D.A, June, C.M, Szarecka, A, Wawrzak, Z. | | Deposit date: | 2023-05-04 | | Release date: | 2024-05-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structural and Dynamic Features of Acinetobacter baumannii OXA-66 beta-Lactamase Explain Its Stability and Evolution of Novel Variants.
J.Mol.Biol., 436, 2024
|
|
4RMH
 
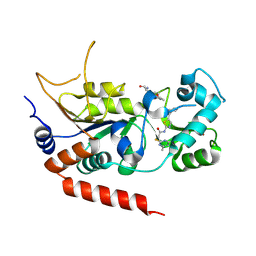 | | Human Sirt2 in complex with SirReal2 and Ac-Lys-H3 peptide | | Descriptor: | 2-[(4,6-dimethylpyrimidin-2-yl)sulfanyl]-N-[5-(naphthalen-1-ylmethyl)-1,3-thiazol-2-yl]acetamide, Ac-Lys-H3 peptide, NAD-dependent protein deacetylase sirtuin-2, ... | | Authors: | Rumpf, T, Schiedel, M, Karaman, B, Roessler, C, North, B.J, Lehotzky, A, Olah, J, Ladwein, K.I, Schmidtkunz, K, Gajer, M, Pannek, M, Steegborn, C, Sinclair, D.A, Gerhardt, S, Ovadi, J, Schutkowski, M, Sippl, W, Einsle, O, Jung, M. | | Deposit date: | 2014-10-21 | | Release date: | 2015-02-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Selective Sirt2 inhibition by ligand-induced rearrangement of the active site.
Nat Commun, 6, 2015
|
|
4RMJ
 
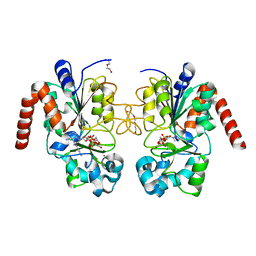 | | Human Sirt2 in complex with ADP ribose and nicotinamide | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, NAD-dependent protein deacetylase sirtuin-2, ... | | Authors: | Rumpf, T, Schiedel, M, Karaman, B, Roessler, C, North, B.J, Lehotzky, A, Olah, J, Ladwein, K.I, Schmidtkunz, K, Gajer, M, Pannek, M, Steegborn, C, Sinclair, D.A, Gerhardt, S, Ovadi, J, Schutkowski, M, Sippl, W, Einsle, O, Jung, M. | | Deposit date: | 2014-10-21 | | Release date: | 2015-02-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Selective Sirt2 inhibition by ligand-induced rearrangement of the active site.
Nat Commun, 6, 2015
|
|
4TLP
 
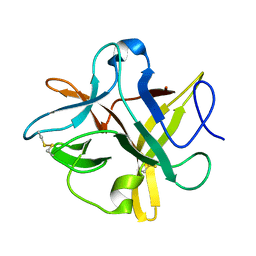 | | Crystal structure of a alanine91 mutant of WCI | | Descriptor: | Chymotrypsin inhibitor 3 | | Authors: | Sen, U, Majumder, S. | | Deposit date: | 2014-05-30 | | Release date: | 2014-07-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A conserved tryptophan (W91) at the barrel-lid junction modulates the packing and stability of Kunitz (STI) family of inhibitors.
Biochim.Biophys.Acta, 1854, 2015
|
|
4RMI
 
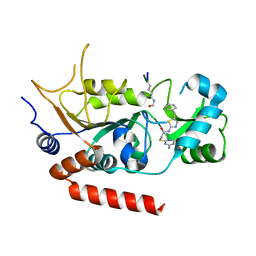 | | Human Sirt2 in complex with SirReal1 and Ac-Lys-OTC peptide | | Descriptor: | Ac-Lys-OTC peptide, N-(5-benzyl-1,3-thiazol-2-yl)-2-[(4,6-dimethylpyrimidin-2-yl)sulfanyl]acetamide, NAD-dependent protein deacetylase sirtuin-2, ... | | Authors: | Rumpf, T, Schiedel, M, Karaman, B, Roessler, C, North, B.J, Lehotzky, A, Olah, J, Ladwein, K.I, Schmidtkunz, K, Gajer, M, Pannek, M, Steegborn, C, Sinclair, D.A, Gerhardt, S, Ovadi, J, Schutkowski, M, Sippl, W, Einsle, O, Jung, M. | | Deposit date: | 2014-10-21 | | Release date: | 2015-02-25 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Selective Sirt2 inhibition by ligand-induced rearrangement of the active site.
Nat Commun, 6, 2015
|
|
8FQW
 
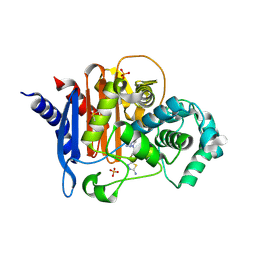 | | ADC-30 in complex with boronic acid transition state inhibitor MB076 | | Descriptor: | 1-[(2R)-2-{2-[(5-amino-1,3,4-thiadiazol-2-yl)sulfanyl]acetamido}-2-boronoethyl]-1H-1,2,3-triazole-4-carboxylic acid, Beta-lactamase, DIMETHYL SULFOXIDE, ... | | Authors: | Powers, R.A, Wallar, B.J, June, C.M, Ruiz, V.J. | | Deposit date: | 2023-01-06 | | Release date: | 2023-07-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Synthesis of a Novel Boronic Acid Transition State Inhibitor, MB076: A Heterocyclic Triazole Effectively Inhibits Acinetobacter -Derived Cephalosporinase Variants with an Expanded-Substrate Spectrum.
J.Med.Chem., 66, 2023
|
|
