6PP4
 
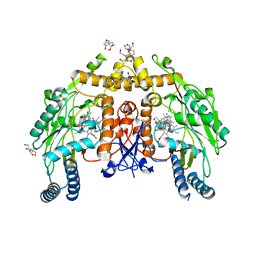 | | Structure of human endothelial nitric oxide synthase heme domain in complex with 7-(3-(Aminomethyl)-4-(pyridin-3-ylmethoxy)phenyl)-4-methylquinolin-2-amine | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5,6,7,8-TETRAHYDROBIOPTERIN, 7-{3-(aminomethyl)-4-[(pyridin-3-yl)methoxy]phenyl}-4-methylquinolin-2-amine, ... | | Authors: | Chreifi, G, Li, H, Poulos, T.L. | | Deposit date: | 2019-07-05 | | Release date: | 2020-04-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | First Contact: 7-Phenyl-2-Aminoquinolines, Potent and Selective Neuronal Nitric Oxide Synthase Inhibitors That Target an Isoform-Specific Aspartate.
J.Med.Chem., 63, 2020
|
|
7O45
 
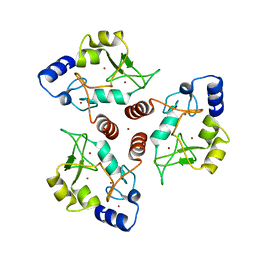 | | Crystal structure of ADD domain of the human DNMT3B methyltransferase | | Descriptor: | BROMIDE ION, Isoform 6 of DNA (cytosine-5)-methyltransferase 3B, ZINC ION | | Authors: | Boyko, K.M, Nikolaeva, A.Y, Bonchuk, A.N, Georgiev, P.G, Popov, V.O. | | Deposit date: | 2021-04-05 | | Release date: | 2022-04-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the DNMT3B ADD domain suggests the absence of a DNMT3A-like autoinhibitory mechanism.
Biochem.Biophys.Res.Commun., 619, 2022
|
|
6UWY
 
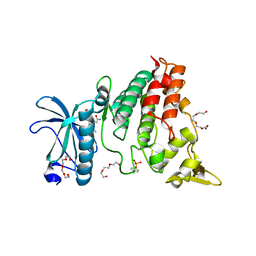 | | DYRK1A bound to a harmine derivative | | Descriptor: | 4-(7-methoxy-1-methyl-9H-beta-carbolin-9-yl)butanamide, Dual specificity tyrosine-phosphorylation-regulated kinase 1A, TETRAETHYLENE GLYCOL | | Authors: | Khamrui, S, Lazarus, M.B. | | Deposit date: | 2019-11-05 | | Release date: | 2020-02-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Synthesis and Biological Validation of a Harmine-Based, Central Nervous System (CNS)-Avoidant, Selective, Human beta-Cell Regenerative Dual-Specificity Tyrosine Phosphorylation-Regulated Kinase A (DYRK1A) Inhibitor.
J.Med.Chem., 63, 2020
|
|
6PC9
 
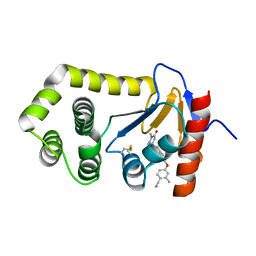 | | Crystal Structure of EcDsbA in a complex with purified methylpiperazinone 6 | | Descriptor: | 2-methyl-4-{4-[2-(4-methyl-3-oxopiperazin-1-yl)-2-oxoethyl]phenoxy}benzonitrile, TRIETHYLENE GLYCOL, Thiol:disulfide interchange protein DsbA | | Authors: | Ilyichova, O.V, Bentley, M, Doak, B, Scanlon, M.J. | | Deposit date: | 2019-06-17 | | Release date: | 2020-05-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Rapid Elaboration of Fragments into Leads by X-ray Crystallographic Screening of Parallel Chemical Libraries (REFiLX).
J.Med.Chem., 63, 2020
|
|
6AXF
 
 | | Structure of RasGRP2 in complex with Rap1B | | Descriptor: | RAS guanyl-releasing protein 2, Ras-related protein Rap-1b | | Authors: | Kondo, Y, Iwig, J.S, Kuriyan, J. | | Deposit date: | 2017-09-06 | | Release date: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | A histidine pH sensor regulates activation of the Ras-specific guanine nucleotide exchange factor RasGRP1.
Elife, 6, 2017
|
|
8Q71
 
 | | Crystal structure of SARS-CoV-2 main protease (MPro) in complex with the inhibitor GC-67 | | Descriptor: | (2~{S})-1-(3,4-dichlorophenyl)-4-(4-methoxypyridin-3-yl)carbonyl-~{N}-(thiophen-2-ylmethyl)piperazine-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Strater, N, Muller, C.E, Sylvester, K, Weisse, R.H, Useini, A, Gao, S, Song, L, Liu, Z, Zhan, P. | | Deposit date: | 2023-08-15 | | Release date: | 2023-12-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.322 Å) | | Cite: | Design, Synthesis, and Biological Evaluation of Trisubstituted Piperazine Derivatives as Noncovalent Severe Acute Respiratory Syndrome Coronavirus 2 Main Protease Inhibitors with Improved Antiviral Activity and Favorable Druggability.
J.Med.Chem., 66, 2023
|
|
6MFY
 
 | | Crystal structure of a 5-domain construct of LgrA in the substrate donation state | | Descriptor: | 4'-PHOSPHOPANTETHEINE, Linear gramicidin synthase subunit A, PHOSPHATE ION | | Authors: | Reimer, J.M, Eivaskhani, M, Harb, I, Schmeing, T.M. | | Deposit date: | 2018-09-12 | | Release date: | 2019-11-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of a dimodular nonribosomal peptide synthetase reveal conformational flexibility.
Science, 366, 2019
|
|
7SOI
 
 | | Structure of I552A Soybean Lipoxygenase at 277K | | Descriptor: | FE (III) ION, Lipoxygenase, SODIUM ION | | Authors: | Gee, C.L, Offenbacher, A.R, Hu, S. | | Deposit date: | 2021-10-31 | | Release date: | 2022-11-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Temporal and spatial resolution of distal protein motions that activate hydrogen tunneling in soybean lipoxygenase.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
6AZO
 
 | | Structural and biochemical characterization of a non-canonical biuret hydrolase (BiuH) from the cyanuric acid catabolism pathway of Rhizobium leguminasorum bv. viciae 3841 | | Descriptor: | CHLORIDE ION, Putative amidase | | Authors: | Peat, T.S, Esquirol, L, Newman, J, Scott, C. | | Deposit date: | 2017-09-11 | | Release date: | 2018-02-21 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Structural and biochemical characterization of the biuret hydrolase (BiuH) from the cyanuric acid catabolism pathway of Rhizobium leguminasorum bv. viciae 3841.
PLoS ONE, 13, 2018
|
|
8DB3
 
 | | Crystal structure of KaiC with truncated C-terminal coiled-coil domain | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Circadian clock protein KaiC | | Authors: | Padua, R.A.P, Grant, T, Pitsawong, W, Hoemberger, M.S, Otten, R, Bradshaw, N, Grigorieff, N, Kern, D. | | Deposit date: | 2022-06-14 | | Release date: | 2023-03-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | From primordial clocks to circadian oscillators.
Nature, 616, 2023
|
|
6PCW
 
 | | Human PIM1 bound to benzothiophene inhibitor 213 | | Descriptor: | 4-[5-(cyclopropylcarbamoyl)thiophen-2-yl]-1-benzothiophene-2-carboxamide, GLYCEROL, Peptide, ... | | Authors: | Godoi, P.H.C, Santiago, A.S, Fala, A.M, Ramos, P.Z, Sriranganadane, D, Mascarello, A, Segretti, N, Azevedo, H, Guimaraes, C.R.W, Arruda, P, Elkins, J.M, Counago, R.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-06-18 | | Release date: | 2019-07-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | PIM1 bound to benzothiophene inhibitor
To Be Published
|
|
6AZT
 
 | |
7T0U
 
 | | Crystal structure of the BCL6 BTB domain in complex with OICR-12387 | | Descriptor: | 3-chloro-5-{7-[2-({5-chloro-2-[(2S)-2-methyl-4-(oxetan-3-yl)piperazin-1-yl]pyridin-4-yl}amino)-2-oxoethyl]-3-methyl-4-oxo-2-(trifluoromethyl)-4,7-dihydro-3H-pyrrolo[2,3-d]pyrimidin-5-yl}-2-hydroxybenzamide, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Kuntz, D.A, Prive, G.G. | | Deposit date: | 2021-11-30 | | Release date: | 2022-11-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Crystal structure of the BCL6 BTB domain in complex with OICR-12387
To Be Published
|
|
6OZL
 
 | |
6MA4
 
 | | Crystal structure of human O-GlcNAc transferase bound to a peptide from HCF-1 pro-repeat 2 (11-26) and inhibitor 3a | | Descriptor: | 5-{2-[(1R)-2-{(carboxymethyl)[(thiophen-2-yl)methyl]amino}-2-oxo-1-{[(2-oxo-1,2-dihydroquinolin-6-yl)sulfonyl]amino}ethyl]phenoxy}pentanoic acid, Host Cell Factor 1 peptide, UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunit | | Authors: | Martin, S.E.S, Lazarus, M.B, Walker, S. | | Deposit date: | 2018-08-25 | | Release date: | 2018-10-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Based Evolution of Low Nanomolar O-GlcNAc Transferase Inhibitors.
J. Am. Chem. Soc., 140, 2018
|
|
7SNS
 
 | | 1.55A Resolution Structure of NanoLuc Luciferase | | Descriptor: | ACETATE ION, Oplophorus-luciferin 2-monooxygenase catalytic subunit | | Authors: | Lovell, S, Mehzabeen, N, Battaile, K.P, Wood, M.G, Encell, L.P, Wood, K.V. | | Deposit date: | 2021-10-28 | | Release date: | 2022-11-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | 1.55A Resolution Structure of NanoLuc Luciferase
To be published
|
|
6VFT
 
 | | Crystal structure of human delta protocadherin 17 EC1-EC4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Harrison, O.J, Brasch, J, Shapiro, L. | | Deposit date: | 2020-01-06 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.71 Å) | | Cite: | Family-wide Structural and Biophysical Analysis of Binding Interactions among Non-clustered delta-Protocadherins.
Cell Rep, 30, 2020
|
|
7O1N
 
 | |
7O3K
 
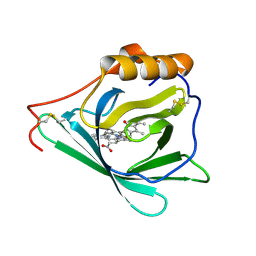 | |
7WM7
 
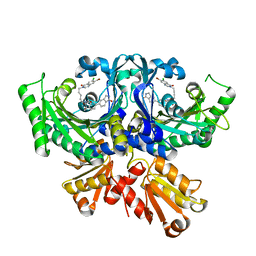 | | Threonyl-tRNA synthetase from Salmonella enterica in complex with an inhibitor | | Descriptor: | (2S,3R)-2-azanyl-N-[5-(7-bromanyl-6-chloranyl-4-oxidanylidene-quinazolin-3-yl)pentyl]-3-oxidanyl-N-[[3-(1-oxidanylidene-2,3-dihydroisoindol-5-yl)phenyl]methyl]butanamide, Threonine--tRNA ligase, ZINC ION | | Authors: | Cai, Z, Chen, B, Yu, Y, Zhou, H. | | Deposit date: | 2022-01-14 | | Release date: | 2022-04-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Design, Synthesis, and Proof-of-Concept of Triple-Site Inhibitors against Aminoacyl-tRNA Synthetases.
J.Med.Chem., 65, 2022
|
|
6P01
 
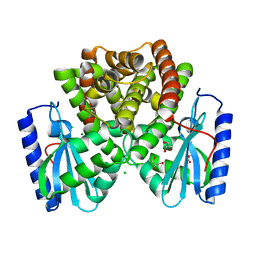 | | Apo structure of the E52D mutant of ANT-4 | | Descriptor: | CHLORIDE ION, GLYCEROL, Kanamycin nucleotidyltransferase, ... | | Authors: | Selvaraj, B, Cuneo, M.J. | | Deposit date: | 2019-05-16 | | Release date: | 2020-01-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | "Catch and Release": a Variation of the Archetypal Nucleotidyl Transfer Reaction
Acs Catalysis, 10, 2020
|
|
6PDV
 
 | | Cu-Carbonic Anhydrase II, A Nitrite Reductase | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, COPPER (II) ION, Carbonic anhydrase 2, ... | | Authors: | Andring, J.T, McKenna, R. | | Deposit date: | 2019-06-19 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Structure and mechanism of copper-carbonic anhydrase II: a nitrite reductase.
Iucrj, 7, 2020
|
|
7SVT
 
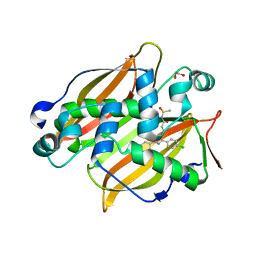 | | Mycobacterium tuberculosis 3-hydroxyl-ACP dehydratase HadAB in complex with 1,3-diarylpyrazolyl-acylsulfonamide inhibitor | | Descriptor: | (3R)-hydroxyacyl-ACP dehydratase subunit HadB, 1,2-ETHANEDIOL, 3-[1-(4-bromophenyl)-3-(4-chlorophenyl)-1H-pyrazol-4-yl]-N-(methanesulfonyl)propanamide, ... | | Authors: | Krieger, I.V, Sacchettini, J.C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2021-11-19 | | Release date: | 2022-11-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | 1,3-Diarylpyrazolyl-acylsulfonamides Target HadAB/BC Complex in Mycobacterium tuberculosis .
Acs Infect Dis., 8, 2022
|
|
6VGO
 
 | |
8DJC
 
 | | CRYSTAL STRUCTURE OF GLYCOGEN SYNTHASE KINASE 3 BETA COMPLEXED WITH (4S)-N-{4-[(2S)-2-methylmorpholin-4-yl] pyridin-3-yl}-2-phenylimidazo[1,2-b]pyridazine-8-carboxamide | | Descriptor: | (4S)-N-{4-[(2S)-2-methylmorpholin-4-yl]pyridin-3-yl}-2-phenylimidazo[1,2-b]pyridazine-8-carboxamide, Glycogen synthase kinase-3 beta | | Authors: | Lewis, H.A, Muckelbauer, J.K. | | Deposit date: | 2022-06-30 | | Release date: | 2023-03-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.463 Å) | | Cite: | Design, Structure-Activity Relationships, and In Vivo Evaluation of Potent and Brain-Penetrant Imidazo[1,2- b ]pyridazines as Glycogen Synthase Kinase-3 beta (GSK-3 beta ) Inhibitors.
J.Med.Chem., 66, 2023
|
|
