7MPU
 
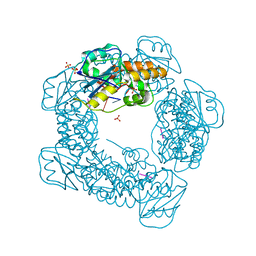 | | Brucella melitensis NrnC bound to pGG | | Descriptor: | 5'-phosphorylated GG, NanoRNase C, SULFATE ION | | Authors: | Lormand, J.D, Sondermann, H. | | Deposit date: | 2021-05-04 | | Release date: | 2021-09-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Structural characterization of NrnC identifies unifying features of dinucleotidases.
Elife, 10, 2021
|
|
7P1N
 
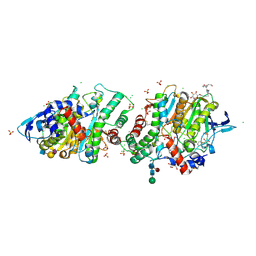 | | Crystal structure of human acetylcholinesterase in complex with (2R,3R,4S,5S,6R)-2-{4-[1-(4-{5-hydroxy-6-[(E)-(hydroxyimino)methyl]pyridin-2-yl}butyl)-1H-1,2,3-triazol-4-yl]butoxy}-6-(hydroxymethyl)oxane-3,4,5-triol oxime | | Descriptor: | (2R,3R,4S,5S,6R)-2-[4-[1-[4-[6-[(Z)-hydroxyiminomethyl]-5-oxidanyl-pyridin-2-yl]butyl]-1,2,3-triazol-4-yl]butoxy]-6-(hydroxymethyl)oxane-3,4,5-triol, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholinesterase, ... | | Authors: | Da Silva, O, Dias, J, Nachon, F. | | Deposit date: | 2021-07-02 | | Release date: | 2022-06-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | A New Class of Bi- and Trifunctional Sugar Oximes as Antidotes against Organophosphorus Poisoning.
J.Med.Chem., 65, 2022
|
|
7MLC
 
 | | PYL10 bound to the ABA pan-antagonist 4a | | Descriptor: | 1-{2-[3,5-dicyclopropyl-4-(4-{[(quinoxaline-2-carbonyl)amino]methyl}-1H-1,2,3-triazol-1-yl)phenyl]acetamido}cyclohexane-1-carboxylic acid, Abscisic acid receptor PYL10, GLYCEROL | | Authors: | Peterson, F.C, Vaidya, A.S, Volkman, B.F, Cutler, S.R. | | Deposit date: | 2021-04-28 | | Release date: | 2021-09-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Click-to-lead design of a picomolar ABA receptor antagonist with potent activity in vivo.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
8GIT
 
 | |
8GIS
 
 | |
7MLD
 
 | | PYL10 bound to the ABA pan-antagonist antabactin | | Descriptor: | Abscisic acid receptor PYL10, CITRIC ACID, antabactin | | Authors: | Peterson, F.C, Vaidya, A.S, Volkman, B.F, Cutler, S.R. | | Deposit date: | 2021-04-28 | | Release date: | 2021-09-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Click-to-lead design of a picomolar ABA receptor antagonist with potent activity in vivo.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
8GIR
 
 | |
8GIO
 
 | |
2ATJ
 
 | | RECOMBINANT HORSERADISH PEROXIDASE COMPLEX WITH BENZHYDROXAMIC ACID | | Descriptor: | BENZHYDROXAMIC ACID, CALCIUM ION, PEROXIDASE C1A, ... | | Authors: | Henriksen, A, Schuller, D.J, Gajhede, M. | | Deposit date: | 1997-08-19 | | Release date: | 1998-01-28 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural interactions between horseradish peroxidase C and the substrate benzhydroxamic acid determined by X-ray crystallography.
Biochemistry, 37, 1998
|
|
8GIM
 
 | |
8GIP
 
 | |
8GIN
 
 | |
8GJR
 
 | |
7OZ8
 
 | |
4V51
 
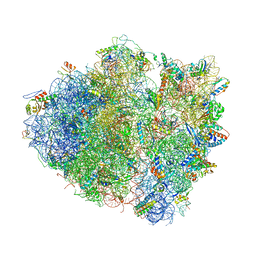 | | Structure of the Thermus thermophilus 70S ribosome complexed with mRNA, tRNA and paromomycin | | Descriptor: | 16S ribosomal RNA, 23S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Selmer, M, Dunham, C.M, Murphy, F.V, Weixlbaumer, A, Petry, S, Weir, J.R, Kelley, A.C, Ramakrishnan, V. | | Deposit date: | 2006-07-31 | | Release date: | 2014-07-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the 70S ribosome complexed with mRNA and tRNA.
Science, 313, 2006
|
|
2AUT
 
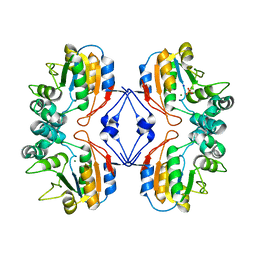 | | Crystal structure of Lys154Asn mutant of mature AphA of S. typhimurium | | Descriptor: | AphA, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Makde, R.D, Gupta, G.D, Kumar, V. | | Deposit date: | 2005-08-29 | | Release date: | 2006-09-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural and mutational analyses reveal the functional role of active-site Lys-154 and Asp-173 of Salmonella typhimurium AphA protein.
Arch.Biochem.Biophys., 464, 2007
|
|
7OZA
 
 | |
8HAD
 
 | |
7OZ9
 
 | |
8HAC
 
 | |
4V41
 
 | | E. COLI (LAC Z) BETA-GALACTOSIDASE (NCS CONSTRAINED MONOMER-MONOCLINIC) | | Descriptor: | BETA-GALACTOSIDASE, MAGNESIUM ION | | Authors: | Juers, D.H, Jacobson, R.H, Wigley, D, Zhang, X.J, Huber, R.E, Tronrud, D.E, Matthews, B.W. | | Deposit date: | 2000-06-07 | | Release date: | 2014-07-09 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | High resolution refinement of beta-galactosidase in a new crystal form reveals multiple metal-binding sites and provides a structural basis for alpha-complementation.
Protein Sci., 9, 2000
|
|
4V6L
 
 | | Structural insights into cognate vs. near-cognate discrimination during decoding. | | Descriptor: | 16S ribosomal RNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Agirrezabala, X, Schreiner, E, Trabuco, L.G, Lei, J, Ortiz-Meoz, R.F, Schulten, K, Green, R, Frank, J. | | Deposit date: | 2011-01-07 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (13.2 Å) | | Cite: | Structural insights into cognate versus near-cognate discrimination during decoding.
Embo J., 30, 2011
|
|
7P19
 
 | |
8H05
 
 | |
2ALC
 
 | | ETHANOL REGULON TRANSCRIPTIONAL ACTIVATOR DNA-BINDING DOMAIN FROM ASPERGILLUS NIDULANS | | Descriptor: | PROTEIN (ETHANOL REGULON TRANSCRIPTIONAL ACTIVATOR), ZINC ION | | Authors: | Cerdan, R, Cahuzac, B, Felenbok, B, Guittet, E. | | Deposit date: | 1999-01-20 | | Release date: | 2000-01-21 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of AlcR (1-60) provides insight in the unusual DNA binding properties of this zinc binuclear cluster protein.
J.Mol.Biol., 295, 2000
|
|
