8D13
 
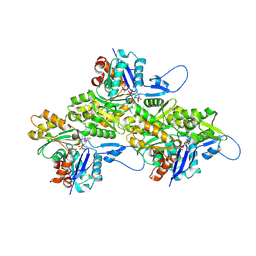 | | Helical ADP-F-actin | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Reynolds, M.J, Alushin, G.M. | | Deposit date: | 2022-05-26 | | Release date: | 2022-09-07 | | Last modified: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (2.43 Å) | | Cite: | Bending forces and nucleotide state jointly regulate F-actin structure.
Nature, 611, 2022
|
|
8DLU
 
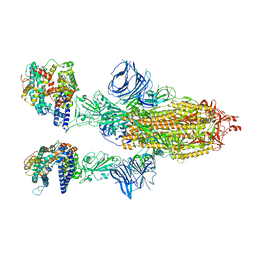 | | Cryo-EM structure of SARS-CoV-2 Epsilon (B.1.429) spike protein in complex with human ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Zhu, X, Mannar, D, Saville, J.W, Srivastava, S.S, Berezuk, A.M, Zhou, S, Tuttle, K.S, Subramaniam, S. | | Deposit date: | 2022-07-08 | | Release date: | 2022-08-31 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | SARS-CoV-2 variants of concern: spike protein mutational analysis and epitope for broad neutralization.
Nat Commun, 13, 2022
|
|
8D1D
 
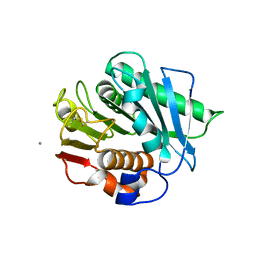 | | PROSS PETase | | Descriptor: | CALCIUM ION, Poly(ethylene terephthalate) hydrolase | | Authors: | Vongsouthi, V, Jackson, C.J, Tan, L. | | Deposit date: | 2022-05-27 | | Release date: | 2022-09-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | PROSS PETase
To Be Published
|
|
8CYB
 
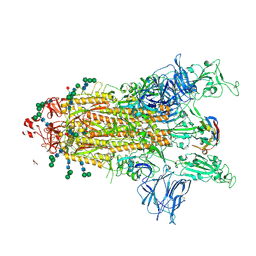 | |
8DEP
 
 | | Cryo-EM structure of the human reduced folate carrier, apo condition | | Descriptor: | Digitonin, Reduced folate transporter,Soluble cytochrome b562 | | Authors: | Wright, N.J, Fedor, J.G, Lee, S.-Y. | | Deposit date: | 2022-06-21 | | Release date: | 2022-09-21 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Methotrexate recognition by the human reduced folate carrier SLC19A1.
Nature, 609, 2022
|
|
5YM7
 
 | |
8DKE
 
 | |
8DKW
 
 | |
8CYJ
 
 | | RBD of SARS-CoV-2 Spike protein in complex with pan-sarbecovirus nanobodies 2-10, 2-67, 2-62 and 1-25 | | Descriptor: | Spike glycoprotein, pan-sarbecovirus nanobody 1-25, pan-sarbecovirus nanobody 2-10, ... | | Authors: | Huang, W, Taylor, D. | | Deposit date: | 2022-05-23 | | Release date: | 2022-07-06 | | Last modified: | 2022-07-13 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Superimmunity by pan-sarbecovirus nanobodies.
Cell Rep, 39, 2022
|
|
8DLM
 
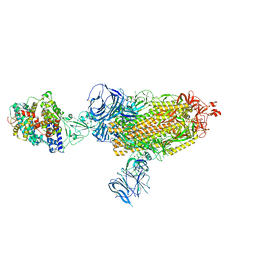 | | Cryo-EM structure of SARS-CoV-2 Beta (B.1.351) spike protein in complex with human ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Zhu, X, Mannar, D, Saville, J.W, Srivastava, S.S, Berezuk, A.M, Zhou, S, Tuttle, K.S, Subramaniam, S. | | Deposit date: | 2022-07-08 | | Release date: | 2022-08-31 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | SARS-CoV-2 variants of concern: spike protein mutational analysis and epitope for broad neutralization.
Nat Commun, 13, 2022
|
|
5EKV
 
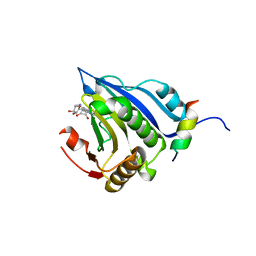 | | Co-crystal structure of eIF4E with nucleotide mimetic inhibitor. | | Descriptor: | 3-[[(2~{R},3~{S},4~{R},5~{R})-5-(2-azanyl-7-methyl-6-oxidanylidene-1~{H}-purin-7-ium-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methylamino]-4-oxidanyl-cyclobut-3-ene-1,2-dione, Eukaryotic translation initiation factor 4E, Eukaryotic translation initiation factor 4E-binding protein 1 | | Authors: | Nowicki, M.W, Walkinshaw, M.D, Fischer, P.M. | | Deposit date: | 2015-11-04 | | Release date: | 2016-09-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.61 Å) | | Cite: | Design of nucleotide-mimetic and non-nucleotide inhibitors of the translation initiation factor eIF4E: Synthesis, structural and functional characterisation.
Eur.J.Med.Chem., 124, 2016
|
|
8D17
 
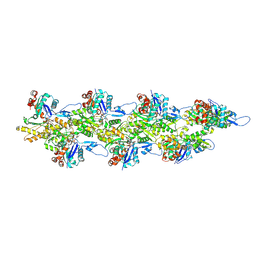 | | Straight ADP-F-actin 1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Reynolds, M.J, Alushin, G.M. | | Deposit date: | 2022-05-26 | | Release date: | 2022-09-07 | | Last modified: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (3.69 Å) | | Cite: | Bending forces and nucleotide state jointly regulate F-actin structure.
Nature, 611, 2022
|
|
8CU7
 
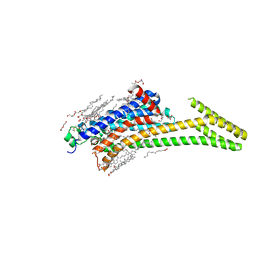 | | Crystal structure of A2AAR-StaR2-bRIL in complex with a novel A2a antagonist, LJ-4517 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2R,3R,4R)-2-[(8P)-6-amino-2-(hex-1-yn-1-yl)-8-(thiophen-2-yl)-9H-purin-9-yl]oxolane-3,4-diol, Adenosine receptor A2a,Soluble cytochrome b562, ... | | Authors: | Shiriaeva, A, Park, D.-J, Kim, G, Lee, Y, Hou, X, Jarhad, D.B, Kim, G, Yu, J, Hyun, Y.E, Kim, W, Gao, Z.-G, Jacobson, K.A, Han, G.W, Stevens, R.C, Jeong, L.S, Choi, S, Cherezov, V. | | Deposit date: | 2022-05-16 | | Release date: | 2022-08-31 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | GPCR Agonist-to-Antagonist Conversion: Enabling the Design of Nucleoside Functional Switches for the A 2A Adenosine Receptor.
J.Med.Chem., 65, 2022
|
|
6PMV
 
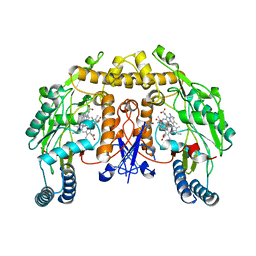 | | Structure of rat neuronal nitric oxide synthase heme domain in complex with 7-(4-(2-Aminoethyl)phenyl)-4-methylquinolin-2-amine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 7-[4-(2-aminoethyl)phenyl]-4-methylquinolin-2-amine, ACETATE ION, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2019-07-02 | | Release date: | 2020-04-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | First Contact: 7-Phenyl-2-Aminoquinolines, Potent and Selective Neuronal Nitric Oxide Synthase Inhibitors That Target an Isoform-Specific Aspartate.
J.Med.Chem., 63, 2020
|
|
6PN6
 
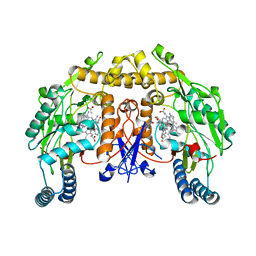 | | Structure of rat neuronal nitric oxide synthase heme domain in complex with 7-(3-(Aminomethyl)-4-(pyridin-3-ylmethoxy)phenyl)-4-methylquinolin-2-amine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 7-{3-(aminomethyl)-4-[(pyridin-3-yl)methoxy]phenyl}-4-methylquinolin-2-amine, ACETATE ION, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2019-07-02 | | Release date: | 2020-04-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.843 Å) | | Cite: | First Contact: 7-Phenyl-2-Aminoquinolines, Potent and Selective Neuronal Nitric Oxide Synthase Inhibitors That Target an Isoform-Specific Aspartate.
J.Med.Chem., 63, 2020
|
|
5YO6
 
 | |
5YO3
 
 | | Crystal Structure of B562RIL with engineered disulfide bond V16C-A29C | | Descriptor: | SULFATE ION, Soluble cytochrome b562 | | Authors: | Pu, M, Xu, Z, Song, G, Liu, Z.J. | | Deposit date: | 2017-10-26 | | Release date: | 2018-05-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Protein crystal quality oriented disulfide bond engineering.
Protein Cell, 9, 2018
|
|
6PBD
 
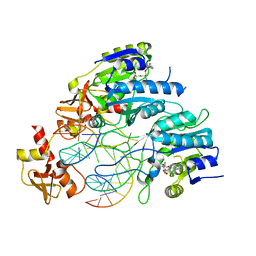 | | DNA N6-Adenine Methyltransferase CcrM In Complex with Double-Stranded DNA Oligonucleotide Containing Its Recognition Sequence GAATC | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*CP*GP*AP*TP*TP*CP*AP*AP*TP*GP*AP*AP*TP*CP*CP*CP*AP*AP*G)-3'), DNA (5'-D(*GP*CP*TP*TP*GP*GP*GP*AP*TP*TP*CP*AP*TP*TP*GP*AP*AP*TP*C)-3'), ... | | Authors: | Horton, J.R, Cheng, X, Woodcock, C.B. | | Deposit date: | 2019-06-13 | | Release date: | 2019-10-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.343 Å) | | Cite: | The cell cycle-regulated DNA adenine methyltransferase CcrM opens a bubble at its DNA recognition site.
Nat Commun, 10, 2019
|
|
8D16
 
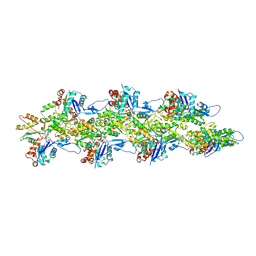 | | Bent ADP-Pi-F-actin | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Reynolds, M.J, Alushin, G.M. | | Deposit date: | 2022-05-26 | | Release date: | 2022-09-07 | | Last modified: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (3.71 Å) | | Cite: | Bending forces and nucleotide state jointly regulate F-actin structure.
Nature, 611, 2022
|
|
5EE7
 
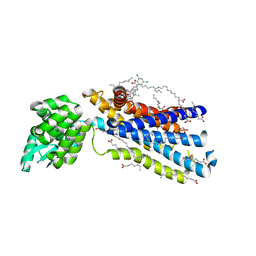 | | Crystal structure of the human glucagon receptor (GCGR) in complex with the antagonist MK-0893 | | Descriptor: | 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, 3-[[4-[(1~{S})-1-[3-[3,5-bis(chloranyl)phenyl]-5-(6-methoxynaphthalen-2-yl)pyrazol-1-yl]ethyl]phenyl]carbonylamino]propanoic acid, Glucagon receptor,Endolysin,Glucagon receptor, ... | | Authors: | Jazayeri, A, Dore, A.S, Lamb, D, Krishnamurthy, H, Southall, S.M, Baig, A.H, Bortolato, A, Koglin, M, Robertson, N.J, Errey, J.C, Andrews, S.P, Brown, A.J.H, Cooke, R.M, Weir, M, Marshall, F.H. | | Deposit date: | 2015-10-22 | | Release date: | 2016-04-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Extra-helical binding site of a glucagon receptor antagonist.
Nature, 533, 2016
|
|
8DKI
 
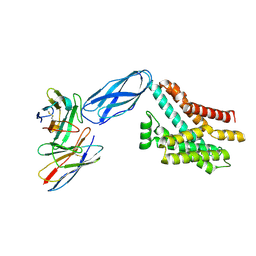 | |
5YPU
 
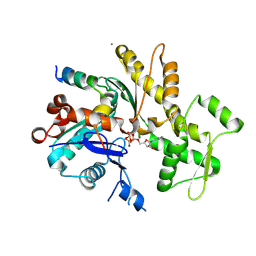 | | Crystal structure of an actin monomer in complex with the nucleator Cordon-Bleu MET72NLE WH2-motif peptide | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Scipion, C.P.M, Wongsantichon, J, Ferrer, F.J, Yuen, T.Y, Robinson, R.C. | | Deposit date: | 2017-11-03 | | Release date: | 2018-09-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural evidence for the roles of divalent cations in actin polymerization and activation of ATP hydrolysis
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5YQ2
 
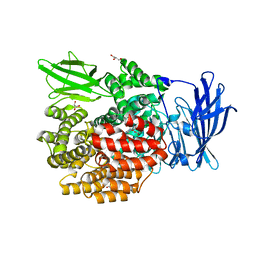 | | Crystal structure of E.coli aminopeptidase N in complex with Puromycin aminonucleoside | | Descriptor: | (2R,3R,4S,5S)-4-AMINO-2-[6-(DIMETHYLAMINO)-9H-PURIN-9-YL]-5-(HYDROXYMETHYL)TETRAHYDRO-3-FURANOL, Aminopeptidase N, GLYCEROL, ... | | Authors: | Marapaka, A.K, Ganji, R.J, Reddi, R, Addlagatta, A. | | Deposit date: | 2017-11-04 | | Release date: | 2018-11-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of E.coli aminopeptidase N in complex with Puromycin aminonucleoside
To Be Published
|
|
8D3Y
 
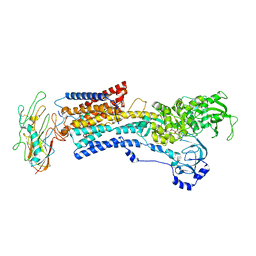 | | Human alpha3 Na+/K+-ATPase in its exoplasmic side-open state | | Descriptor: | FXYD domain-containing ion transport regulator 6, Sodium/potassium-transporting ATPase subunit alpha-3, Sodium/potassium-transporting ATPase subunit beta-1 | | Authors: | Nguyen, P.T, Bai, X. | | Deposit date: | 2022-06-01 | | Release date: | 2022-09-21 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural basis for gating mechanism of the human sodium-potassium pump.
Nat Commun, 13, 2022
|
|
6PJ4
 
 | |
