2NXQ
 
 | | Crystal structure of calcium binding protein 1 from Entamoeba histolytica: a novel arrangement of EF hand motifs | | Descriptor: | ACETATE ION, CALCIUM ION, Calcium-binding protein | | Authors: | Kumar, S, Padhan, N, Alam, N, Gourinath, S. | | Deposit date: | 2006-11-18 | | Release date: | 2007-08-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of calcium binding protein-1 from Entamoeba histolytica: A novel arrangement of EF hand motifs.
Proteins, 68, 2007
|
|
3TTT
 
 | | Structure of F413Y variant of E. coli KatE | | Descriptor: | Catalase HPII, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Loewen, P.C, Jha, V. | | Deposit date: | 2011-09-15 | | Release date: | 2011-10-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Mutation of Phe413 to Tyr in catalase KatE from Escherichia coli leads to side chain damage and main chain cleavage.
Arch.Biochem.Biophys., 525, 2012
|
|
2NXY
 
 | | HIV-1 gp120 Envelope Glycoprotein(S334A) Complexed with CD4 and Antibody 17b | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ANTIBODY 17B, ... | | Authors: | Zhou, T, Xu, L, Dey, B, Hessell, A.J, Van Ryk, D, Xiang, S.H, Yang, X, Zhang, M.Y, Zwick, M.B, Arthos, J, Burton, D.R, Dimitrov, D.S, Sodroski, J, Wyatt, R, Nabel, G.J, Kwong, P.D. | | Deposit date: | 2006-11-20 | | Release date: | 2007-02-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural definition of a conserved neutralization epitope on HIV-1 gp120.
Nature, 445, 2007
|
|
3EM4
 
 | | Crystal structure of atazanavir (ATV) in complex with I50L/A71V drug-resistant HIV-1 protease | | Descriptor: | (3S,8S,9S,12S)-3,12-BIS(1,1-DIMETHYLETHYL)-8-HYDROXY-4,11-DIOXO-9-(PHENYLMETHYL)-6-[[4-(2-PYRIDINYL)PHENYL]METHYL]-2,5, 6,10,13-PENTAAZATETRADECANEDIOIC ACID DIMETHYL ESTER, PHOSPHATE ION, ... | | Authors: | Prabu-Jeyabalan, M, King, N, Royer, C, Schiffer, C. | | Deposit date: | 2008-09-23 | | Release date: | 2009-09-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Kinetic and Structural studies on atazanavir-specific I50L drug-resistant HIV-1 protease mutant
To be Published
|
|
2NVA
 
 | | The X-ray crystal structure of the Paramecium bursaria Chlorella virus arginine decarboxylase bound to agmatine | | Descriptor: | (4-{[(4-{[AMINO(IMINO)METHYL]AMINO}BUTYL)AMINO]METHYL}-5-HYDROXY-6-METHYLPYRIDIN-3-YL)METHYL DIHYDROGEN PHOSPHATE, arginine decarboxylase, A207R protein | | Authors: | Shah, R.H, Akella, R, Goldsmith, E, Phillips, M.A. | | Deposit date: | 2006-11-11 | | Release date: | 2007-03-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray Structure of Paramecium bursaria Chlorella Virus Arginine Decarboxylase: Insight into the Structural Basis for Substrate Specificity.
Biochemistry, 46, 2007
|
|
2NYV
 
 | | X-ray crystal structure of a phosphoglycolate phosphatase from Aquifex aeolicus | | Descriptor: | Phosphoglycolate phosphatase | | Authors: | Ciatto, C, Min, T, Gorman, J, Burley, S.K, Shapiro, L, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-11-21 | | Release date: | 2006-12-05 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.103 Å) | | Cite: | X-ray crystal structure of a phosphoglycolate phosphatase from Aquifex aeolicus
To be Published
|
|
3EOT
 
 | |
3EP6
 
 | | Human AdoMetDC D174N mutant complexed with S-Adenosylmethionine methyl ester and no putrescine bound | | Descriptor: | PYRUVIC ACID, S-ADENOSYLMETHIONINE METHYL ESTER, S-adenosylmethionine decarboxylase alpha chain, ... | | Authors: | Bale, S, Lopez, M.M, Makhatadze, G.I, Fang, Q, Pegg, A.E, Ealick, S.E. | | Deposit date: | 2008-09-29 | | Release date: | 2008-12-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis for Putrescine Activation of Human S-Adenosylmethionine Decarboxylase.
Biochemistry, 47, 2008
|
|
3TZV
 
 | | Crystal structure of an iNKT TCR in complex with CD1d-lysophosphatidylcholine | | Descriptor: | (4R,7R,18E)-4,7-dihydroxy-N,N,N-trimethyl-10-oxo-3,5,9-trioxa-4-phosphaheptacos-18-en-1-aminium 4-oxide, Antigen-presenting glycoprotein CD1d, Beta-2-microglobulin, ... | | Authors: | Lopez-Sagaseta, J, Adams, E.J. | | Deposit date: | 2011-09-27 | | Release date: | 2012-04-25 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.056 Å) | | Cite: | Lysophospholipid presentation by CD1d and recognition by a human Natural Killer T-cell receptor.
Embo J., 31, 2012
|
|
2O3X
 
 | | Crystal Structure of the Prokaryotic Ribosomal Decoding Site Complexed with Paromamine Derivative NB30 | | Descriptor: | (1R,2R,3S,4R,6S)-4,6-DIAMINO-2-[(5-AMINO-5-DEOXY-BETA-D-RIBOFURANOSYL)OXY]-3-HYDROXYCYCLOHEXYL 2-AMINO-2-DEOXY-ALPHA-D-GLUCOPYRANOSIDE, RNA (5'-R(*UP*UP*GP*CP*GP*UP*CP*AP*CP*AP*CP*CP*GP*GP*UP*GP*AP*AP*GP*UP*CP*GP*C)-3') | | Authors: | Kondo, J, Hainrichson, M, Nudelman, I, Shallom-Shezifi, D, Baasov, T, Westhof, E. | | Deposit date: | 2006-12-02 | | Release date: | 2007-11-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Differential Selectivity of Natural and Synthetic Aminoglycosides towards the Eukaryotic and Prokaryotic Decoding A Sites.
Chembiochem, 8, 2007
|
|
3TX6
 
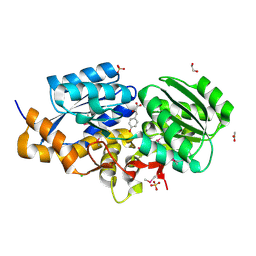 | | The Structure of a putative ABC-transporter periplasmic component from Rhodopseudomonas palustris | | Descriptor: | 1,2-ETHANEDIOL, 3-(4-HYDROXY-PHENYL)PYRUVIC ACID, ACETIC ACID, ... | | Authors: | Cuff, M.E, Mack, J.C, Zerbs, S, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-09-22 | | Release date: | 2011-11-09 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural and functional characterization of solute binding proteins for aromatic compounds derived from lignin: p-Coumaric acid and related aromatic acids.
Proteins, 81, 2013
|
|
3EDY
 
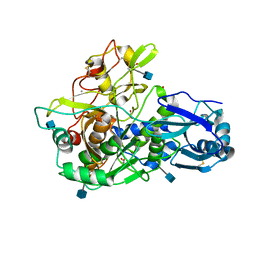 | | Crystal Structure of the Precursor Form of Human Tripeptidyl-Peptidase 1 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Guhaniyogi, J, Sohar, I, Das, K, Lobel, P, Stock, A.M. | | Deposit date: | 2008-09-03 | | Release date: | 2008-11-18 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure and Autoactivation Pathway of the Precursor Form of Human Tripeptidyl-peptidase 1, the Enzyme Deficient in Late Infantile Ceroid Lipofuscinosis
J.Biol.Chem., 284, 2009
|
|
2O36
 
 | |
2O7N
 
 | | CD11A (LFA1) I-domain complexed with 7A-[(4-cyanophenyl)methyl]-6-(3,5-dichlorophenyl)-5-oxo-2,3,5,7A-tetrahydro-1H-pyrrolo[1,2-A]pyrrole-7-carbonitrile | | Descriptor: | 7A-[(4-cyanophenyl)methyl]-6-(3,5-dichlorophenyl)-5-oxo-2,3,5,7A-tetrahydro-1H-pyrrolo[1,2-A]pyrrole-7-carbonitrile, Integrin alpha-L | | Authors: | Sheriff, S. | | Deposit date: | 2006-12-11 | | Release date: | 2007-03-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Design of LFA-1 antagonists based on a 2,3-dihydro-1H-pyrrolizin-5(7aH)-one scaffold.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
3EE7
 
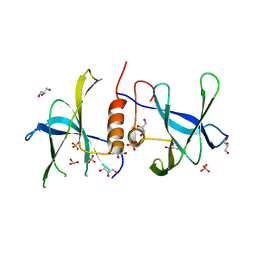 | | Crystal Structure of SARS-CoV nsp9 G104E | | Descriptor: | GLYCEROL, PHOSPHATE ION, Replicase polyprotein 1a | | Authors: | Miknis, Z.J, Donaldson, E.F, Umland, T.C, Rimmer, R, Baric, R.S, Schultz, L.W. | | Deposit date: | 2008-09-04 | | Release date: | 2009-03-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Severe acute respiratory syndrome coronavirus nsp9 dimerization is essential for efficient viral growth
J.Virol., 83, 2009
|
|
2Q0L
 
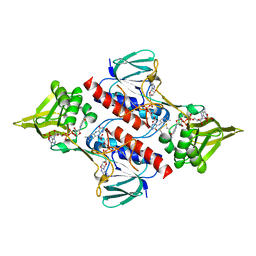 | | Helicobacter pylori thioredoxin reductase reduced by sodium dithionite in complex with NADP+ | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Thioredoxin reductase | | Authors: | Sandalova, T, Gustafsson, T, Lu, J, Holmgren, A, Schneider, G. | | Deposit date: | 2007-05-22 | | Release date: | 2007-07-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | High-resolution structures of oxidized and reduced thioredoxin reductase from Helicobacter pylori.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
3U6I
 
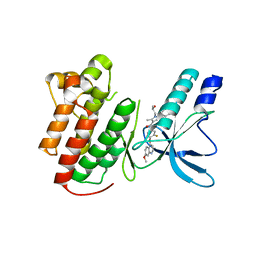 | | Crystal structure of c-Met in complex with pyrazolone inhibitor 58a | | Descriptor: | Hepatocyte growth factor receptor, N-{3-fluoro-4-[(7-methoxyquinolin-4-yl)oxy]phenyl}-1-[(2R)-2-hydroxypropyl]-5-methyl-3-oxo-2-phenyl-2,3-dihydro-1H-pyrazole-4-carboxamide | | Authors: | Bellon, S.F, Whittington, D.A, Long, A.L. | | Deposit date: | 2011-10-12 | | Release date: | 2012-02-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-based design of novel class II c-Met inhibitors: 1. Identification of pyrazolone-based derivatives.
J.Med.Chem., 55, 2012
|
|
3EI1
 
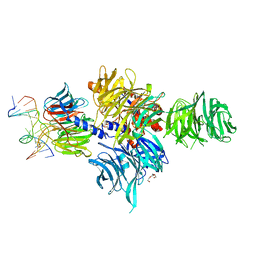 | |
3EIN
 
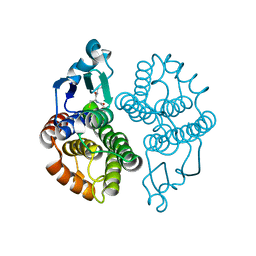 | | Delta class GST | | Descriptor: | GLUTATHIONE, Glutathione S-transferase 1-1 | | Authors: | Feil, S.C. | | Deposit date: | 2008-09-17 | | Release date: | 2009-09-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.126 Å) | | Cite: | Probing insect detoxification systems
To be Published
|
|
2Q31
 
 | | Actin Dimer Cross-linked Between Residues 41 and 374 and proteolytically cleaved by subtilisin between residues 47 and 48. | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Sawaya, M.R, Pashkov, I, Kudryashov, D.S, Reisler, E, Yeates, T.O. | | Deposit date: | 2007-05-29 | | Release date: | 2007-06-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Multiple crystal structures of actin dimers and their implications for interactions in the actin filament.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
3U9W
 
 | | Structure of human Leukotriene A4 hydrolase in complex with inhibitor sc57461A | | Descriptor: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Niegowski, D, Thunnissen, M, Tholander, F, Rinaldo-Matthis, A, Muroya, A, Haeggstrom, J.Z. | | Deposit date: | 2011-10-20 | | Release date: | 2012-10-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structure of human Leukotriene A4 hydrolase in complex with inhibitor sc57461A
To be Published
|
|
3EKU
 
 | | Crystal Structure of Monomeric Actin bound to Cytochalasin D | | Descriptor: | (3S,3aR,4S,6S,6aR,7E,10S,12R,13E,15R,15aR)-3-benzyl-6,12-dihydroxy-4,10,12-trimethyl-5-methylidene-1,11-dioxo-2,3,3a,4,5,6,6a,9,10,11,12,15-dodecahydro-1H-cycloundeca[d]isoindol-15-yl acetate, ADENOSINE-5'-TRIPHOSPHATE, Actin-5C, ... | | Authors: | Nair, U.B, Joel, P.B, Wan, Q, Lowey, S, Rould, M.A, Trybus, K.M. | | Deposit date: | 2008-09-19 | | Release date: | 2008-10-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of monomeric actin bound to cytochalasin D.
J.Mol.Biol., 384, 2008
|
|
2Q5S
 
 | |
3UAZ
 
 | | Crystal structure of Bacillus cereus adenosine phosphorylase D204N mutant complexed with inosine | | Descriptor: | GLYCEROL, INOSINE, Purine nucleoside phosphorylase deoD-type, ... | | Authors: | Dessanti, P, Zhang, Y, Allegrini, S, Tozzi, M.G, Sgarrella, F, Ealick, S.E. | | Deposit date: | 2011-10-22 | | Release date: | 2012-02-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural basis of the substrate specificity of Bacillus cereus adenosine phosphorylase.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3EIV
 
 | |
