8BLP
 
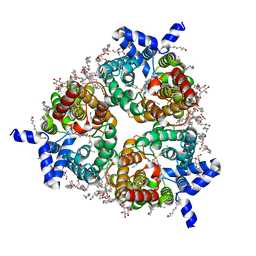 | | Human Urea Transporter UT-B/UT1 in Complex with Inhibitor UTBinh-14 | | Descriptor: | 10-(4-ethylphenyl)sulfonyl-~{N}-(thiophen-2-ylmethyl)-5-thia-1,8,11,12-tetrazatricyclo[7.3.0.0^{2,6}]dodeca-2(6),3,7,9,11-pentaen-7-amine, CHOLESTEROL HEMISUCCINATE, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Chi, G, Dietz, L, Pike, A.C.W, Maclean, E.M, Mukhopadhyay, S.M.M, Bohstedt, T, Wang, D, Scacioc, A, McKinley, G, Arrowsmith, C.H, Edwards, A, Bountra, C, Fernandez-Cid, A, Burgess-Brown, N.A, Duerr, K.L. | | Deposit date: | 2022-11-10 | | Release date: | 2023-10-04 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural characterization of human urea transporters UT-A and UT-B and their inhibition.
Sci Adv, 9, 2023
|
|
7N64
 
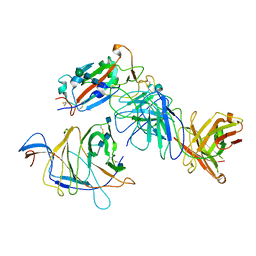 | | SARS-CoV-2 Spike (2P) in complex with G32R7 Fab (RBD and NTD local reconstruction) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, G32R7 Fab heavy chain, ... | | Authors: | Windsor, I.W, Jenni, S, Tong, P, Gautam, A.K, Wesemann, D.R, Harrison, S.C. | | Deposit date: | 2021-06-07 | | Release date: | 2021-08-04 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Memory B cell repertoire for recognition of evolving SARS-CoV-2 spike.
Biorxiv, 2021
|
|
7N62
 
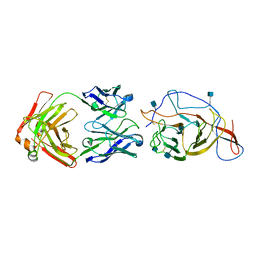 | | SARS-CoV-2 Spike (2P) in complex with C12C9 Fab (NTD local reconstruction) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, C12C9 Fab heavy chain, C12C9 Fab light chain, ... | | Authors: | Windsor, I.W, Jenni, S, Bajic, G, Tong, P, Gautam, A.K, Wesemann, D.R, Harrison, S.C. | | Deposit date: | 2021-06-07 | | Release date: | 2021-08-04 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Memory B cell repertoire for recognition of evolving SARS-CoV-2 spike.
Biorxiv, 2021
|
|
3VD4
 
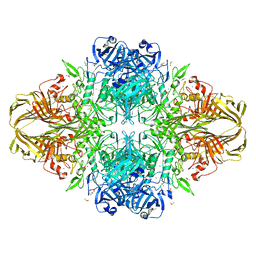 | | E. coli (lacZ) beta-galactosidase (N460D) in complex with IPTG | | Descriptor: | 1-methylethyl 1-thio-beta-D-galactopyranoside, Beta-galactosidase, DIMETHYL SULFOXIDE, ... | | Authors: | Wheatley, R.W, Kappelhoff, J.C, Hahn, J.N, Dugdale, M.L, Dutkoski, M.J, Tamman, S.D, Fraser, M.E, Huber, R.E. | | Deposit date: | 2012-01-04 | | Release date: | 2012-04-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Substitution for Asn460 cripples {beta}-galactosidase (Escherichia coli) by increasing substrate affinity and decreasing transition state stability.
Arch.Biochem.Biophys., 521, 2012
|
|
7NP4
 
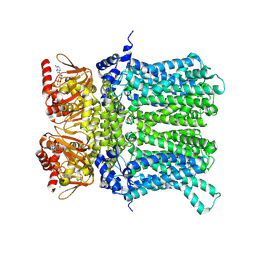 | | cAMP-bound rabbit HCN4 stabilized in LMNG-CHS detergent mixture | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4,Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4 | | Authors: | Giese, H, Chaves-Sanjuan, A, Saponaro, A, Clarke, O, Bolognesi, M, Mancia, F, Hendrickson, W.A, Thiel, G, Santoro, B, Moroni, A. | | Deposit date: | 2021-02-26 | | Release date: | 2021-08-11 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Gating movements and ion permeation in HCN4 pacemaker channels.
Mol.Cell, 81, 2021
|
|
7NP3
 
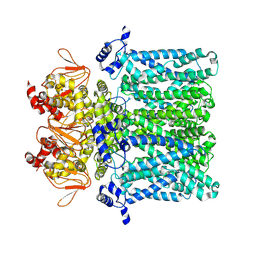 | | cAMP-free rabbit HCN4 stabilized in LMNG-CHS detergent mixture | | Descriptor: | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4,Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4 | | Authors: | Giese, H.M, Chaves-Sanjuan, A, Saponaro, A, Clarke, O, Bolognesi, M, Mancia, F, Hendrickson, W.A, Thiel, G, Santoro, B, Moroni, A. | | Deposit date: | 2021-02-26 | | Release date: | 2021-08-11 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Gating movements and ion permeation in HCN4 pacemaker channels.
Mol.Cell, 81, 2021
|
|
3VI0
 
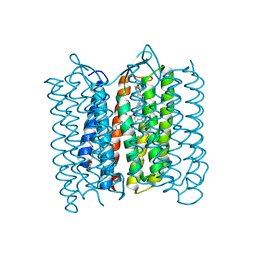 | |
7NIU
 
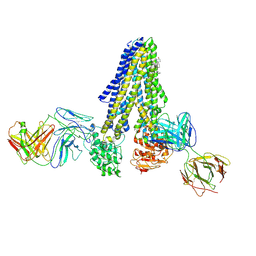 | |
7NIV
 
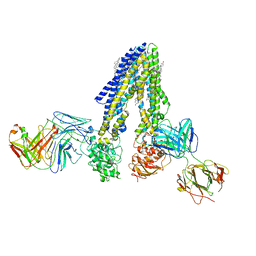 | | Nanodisc reconstituted human ABCB4 in complex with 4B1-Fab and QA2-Fab (phosphatidylcholine-bound, occluded conformation) | | Descriptor: | 1,2-DILINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 4B1 Fab-fragment light chain, CHOLESTEROL, ... | | Authors: | Nosol, K, Locher, K.P. | | Deposit date: | 2021-02-14 | | Release date: | 2021-08-25 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structures of ABCB4 provide insight into phosphatidylcholine translocation.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
8BEJ
 
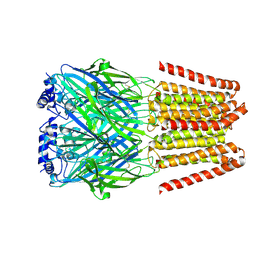 | | GABA-A receptor a5 homomer - a5V3 - APO | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Gamma-aminobutyric acid receptor subunit alpha-5 | | Authors: | Miller, P.S, Malinauskas, T.M, Hardwick, S.W, Chirgadze, D.Y. | | Deposit date: | 2022-10-21 | | Release date: | 2023-11-01 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | The molecular basis of drug selectivity for alpha 5 subunit-containing GABA A receptors.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8BHM
 
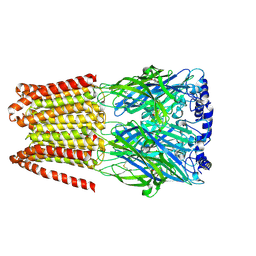 | | GABA-A receptor a5 homomer - a5V3 - DMCM | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Gamma-aminobutyric acid receptor subunit alpha-5, methyl 4-ethyl-6,7-dimethoxy-9H-pyrido[3,4-b]indole-3-carboxylate | | Authors: | Miller, P.S, Malinauskas, T.M, Hardwick, S.W, Chirgadze, D.Y. | | Deposit date: | 2022-10-31 | | Release date: | 2023-11-01 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | The molecular basis of drug selectivity for alpha 5 subunit-containing GABA A receptors.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7N4X
 
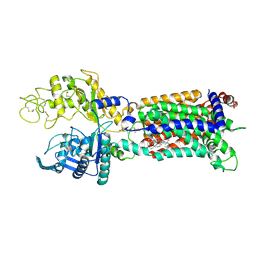 | | Structure of human NPC1L1 mutant-W347R | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Li, X, Long, T. | | Deposit date: | 2021-06-04 | | Release date: | 2021-09-01 | | Last modified: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | Structures of dimeric human NPC1L1 provide insight into mechanisms for cholesterol absorption.
Sci Adv, 7, 2021
|
|
7NIW
 
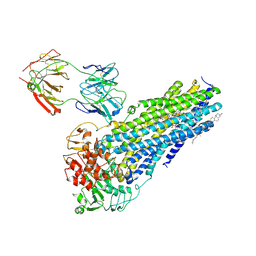 | | Nanodisc reconstituted human ABCB4 in complex with 4B1-Fab (posaconazole-bound, inward-open conformation) | | Descriptor: | 1,2-DILINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 4B1 Fab-fragment heavy chain, 4B1 Fab-fragment light chain, ... | | Authors: | Nosol, K, Locher, K.P. | | Deposit date: | 2021-02-14 | | Release date: | 2021-08-25 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structures of ABCB4 provide insight into phosphatidylcholine translocation.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7N4U
 
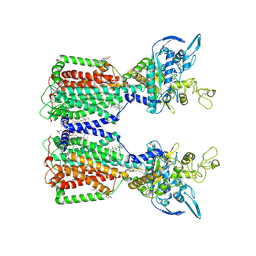 | | Structure of human NPC1L1 | | Descriptor: | (2R)-2,5,7,8-TETRAMETHYL-2-[(4R,8R)-4,8,12-TRIMETHYLTRIDECYL]CHROMAN-6-OL, (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Li, X, Long, T. | | Deposit date: | 2021-06-04 | | Release date: | 2021-09-01 | | Last modified: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (3.34 Å) | | Cite: | Structures of dimeric human NPC1L1 provide insight into mechanisms for cholesterol absorption.
Sci Adv, 7, 2021
|
|
7MYH
 
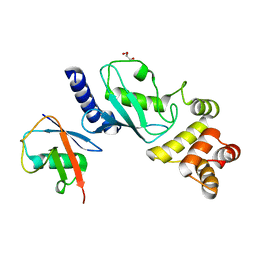 | | Ubiquitin variant UbV.k.2 in complex with Ube2k | | Descriptor: | GLYCEROL, Ubiquitin variant UbV.k.2, Ubiquitin-conjugating enzyme E2 K | | Authors: | Middleton, A.J, Day, C.L, Teyra, J, Sidhu, S.S. | | Deposit date: | 2021-05-21 | | Release date: | 2021-08-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.394 Å) | | Cite: | Identification of Ubiquitin Variants That Inhibit the E2 Ubiquitin Conjugating Enzyme, Ube2k.
Acs Chem.Biol., 16, 2021
|
|
7N4V
 
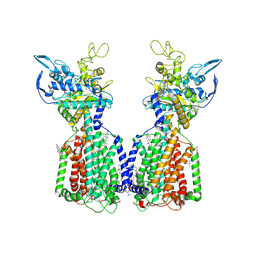 | | Structure of cholesterol-bound human NPC1L1 | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Li, X, Long, T. | | Deposit date: | 2021-06-04 | | Release date: | 2021-09-01 | | Last modified: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | Structures of dimeric human NPC1L1 provide insight into mechanisms for cholesterol absorption.
Sci Adv, 7, 2021
|
|
7N9Z
 
 | | E. coli cytochrome bo3 in MSP nanodisc | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, CARDIOLIPIN, ... | | Authors: | Vallese, F, Clarke, O.B. | | Deposit date: | 2021-06-19 | | Release date: | 2021-09-01 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.19 Å) | | Cite: | Cryo-EM structures of Escherichia coli cytochrome bo 3 reveal bound phospholipids and ubiquinone-8 in a dynamic substrate binding site.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7N7P
 
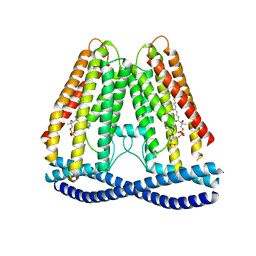 | | Cryo-EM structure of human TMEM120A | | Descriptor: | COENZYME A, Ion channel TACAN | | Authors: | Xue, J, Han, Y, Jiang, Y. | | Deposit date: | 2021-06-10 | | Release date: | 2021-09-01 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | TMEM120A is a coenzyme A-binding membrane protein with structural similarities to ELOVL fatty acid elongase.
Elife, 10, 2021
|
|
8BHB
 
 | | GABA-A receptor a5 homomer - a5V3 - RO154513 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Gamma-aminobutyric acid receptor subunit alpha-5, ethyl 8-[(azanylidene-$l^{4}-azanylidene)amino]-5-methyl-6-oxidanylidene-4~{H}-imidazo[1,5-a][1,4]benzodiazepine-3-carboxylate | | Authors: | Miller, P.S, Malinauskas, T.M, Hardwick, S.W, Chirgadze, D.Y, Wahid, A.A. | | Deposit date: | 2022-10-30 | | Release date: | 2023-11-01 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (2.54 Å) | | Cite: | The molecular basis of drug selectivity for alpha 5 subunit-containing GABA A receptors.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8BHI
 
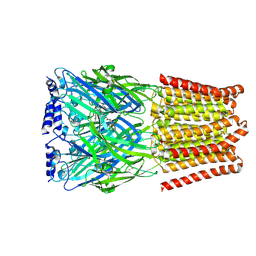 | | GABA-A receptor a5 homomer - a5V3 - RO5211223 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Gamma-aminobutyric acid receptor subunit alpha-5, [1,1-bis(oxidanylidene)-1,4-thiazinan-4-yl]-[6-[[5-methyl-3-(6-methylpyridin-3-yl)-1,2-oxazol-4-yl]methoxy]pyridin-3-yl]methanone | | Authors: | Miller, P.S, Malinauskas, T.M, Hardwick, S.W, Chirgadze, D.Y. | | Deposit date: | 2022-10-31 | | Release date: | 2023-11-01 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (2.67 Å) | | Cite: | The molecular basis of drug selectivity for alpha 5 subunit-containing GABA A receptors.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8BHK
 
 | | GABA-A receptor a5 homomer - a5V3 - Diazepam | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 7-CHLORO-1-METHYL-5-PHENYL-1,3-DIHYDRO-2H-1,4-BENZODIAZEPIN-2-ONE, Gamma-aminobutyric acid receptor subunit alpha-5 | | Authors: | Miller, P.S, Malinauskas, T.M, Kasaragod, V.B, Chirgadze, D.Y. | | Deposit date: | 2022-10-31 | | Release date: | 2023-11-01 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | The molecular basis of drug selectivity for alpha 5 subunit-containing GABA A receptors.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7MYZ
 
 | |
8BHO
 
 | | GABA-A receptor a5 homomer - a5V3 - L655708 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Gamma-aminobutyric acid receptor subunit alpha-5, ethyl (7~{S})-15-methoxy-12-oxidanylidene-2,4,11-triazatetracyclo[11.4.0.0^{2,6}.0^{7,11}]heptadeca-1(17),3,5,13,15-pentaene-5-carboxylate | | Authors: | Miller, P.S, Malinauskas, T.M, Hardwick, S.W, Chirgadze, D.Y. | | Deposit date: | 2022-10-31 | | Release date: | 2023-11-01 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | The molecular basis of drug selectivity for alpha 5 subunit-containing GABA A receptors.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8BHQ
 
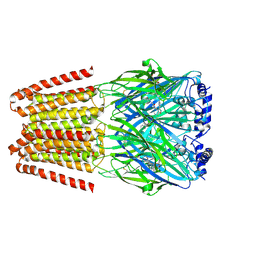 | | GABA-A receptor a5 homomer - a5V3 - RO7172670 | | Descriptor: | 2-[[5-methyl-3-(6-methylpyridazin-3-yl)-1,2-oxazol-4-yl]methyl]-5-(5-oxa-2-azaspiro[3.5]nonan-2-yl)pyridazin-3-one, 2-acetamido-2-deoxy-beta-D-glucopyranose, Gamma-aminobutyric acid receptor subunit alpha-5 | | Authors: | Miller, P.S, Malinauskas, T.M, Hardwick, S.W, Kasaragod, V.B. | | Deposit date: | 2022-10-31 | | Release date: | 2023-11-01 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | The molecular basis of drug selectivity for alpha 5 subunit-containing GABA A receptors.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8BHS
 
 | | GABA-A receptor a5 homomer - a5V3 - RO4938581 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-[bis(fluoranyl)methyl]-15-bromanyl-2,4,8,9,11-pentazatetracyclo[11.4.0.0^{2,6}.0^{8,12}]heptadeca-1(13),3,5,9,11,14,16-heptaene, Gamma-aminobutyric acid receptor subunit alpha-5 | | Authors: | Miller, P.S, Malinauskas, T.M, Hardwick, S.W, Chirgadze, D.Y. | | Deposit date: | 2022-10-31 | | Release date: | 2023-11-01 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | The molecular basis of drug selectivity for alpha 5 subunit-containing GABA A receptors.
Nat.Struct.Mol.Biol., 30, 2023
|
|
