1CCI
 
 | | HOW FLEXIBLE ARE PROTEINS? TRAPPING OF A FLEXIBLE LOOP | | Descriptor: | 2,3-DIMETHYLIMIDAZOLIUM ION, CYTOCHROME C PEROXIDASE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Cao, Y, Musah, R.A, Wilcox, S.K, Goodin, D.B, Mcree, D.E. | | Deposit date: | 1996-12-18 | | Release date: | 1997-07-07 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A ligand-gated, hinged loop rearrangement opens a channel to a buried artificial protein cavity.
Nat.Struct.Biol., 3, 1996
|
|
1CCE
 
 | | CONSTRUCTION OF A BIS-AQUO HEME ENZYME AND REPLACEMENT WITH EXOGENOUS LIGAND | | Descriptor: | CYTOCHROME C PEROXIDASE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Mcree, D.E, Jensen, G.M, Fitzgerald, M.M, Siegel, H.A, Goodin, D.B. | | Deposit date: | 1994-05-04 | | Release date: | 1994-07-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Construction of a bisaquo heme enzyme and binding by exogenous ligands.
Proc.Natl.Acad.Sci.USA, 91, 1994
|
|
1CDE
 
 | | STRUCTURES OF APO AND COMPLEXED ESCHERICHIA COLI GLYCINAMIDE RIBONUCLEOTIDE TRANSFORMYLASE | | Descriptor: | 5-DEAZAFOLIC ACID, GLYCINAMIDE RIBONUCLEOTIDE, PHOSPHORIBOSYL-GLYCINAMIDE FORMYLTRANSFERASE | | Authors: | Almassy, R.J, Janson, C.A, Kan, C.-C, Hostomska, Z. | | Deposit date: | 1992-05-15 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of apo and complexed Escherichia coli glycinamide ribonucleotide transformylase.
Proc.Natl.Acad.Sci.USA, 89, 1992
|
|
1CC6
 
 | | PHE161 AND ARG166 VARIANTS OF P-HYDROXYBENZOATE HYDROXYLASE. IMPLICATIONS FOR NADPH RECOGNITION AND STRUCTURAL STABILITY. | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, P-HYDROXYBENZOIC ACID, PROTEIN (P-HYDROXYBENZOATE HYDROXYLASE) | | Authors: | Eppink, M.H.M, Bunthof, C, Schreuder, H.A, Van Berkel, W.J.H. | | Deposit date: | 1999-03-04 | | Release date: | 1999-03-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Phe161 and Arg166 variants of p-hydroxybenzoate hydroxylase. Implications for NADPH recognition and structural stability.
Febs Lett., 443, 1999
|
|
1CCR
 
 | | STRUCTURE OF RICE FERRICYTOCHROME C AT 2.0 ANGSTROMS RESOLUTION | | Descriptor: | CYTOCHROME C, HEME C | | Authors: | Ochi, H, Hata, Y, Tanaka, N, Kakudo, M, Sakurai, T, Aihara, S, Morita, Y. | | Deposit date: | 1983-03-14 | | Release date: | 1983-04-21 | | Last modified: | 2021-03-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of rice ferricytochrome c at 2.0 A resolution.
J.Mol.Biol., 166, 1983
|
|
1C6X
 
 | |
1C6Z
 
 | | ALTERNATE BINDING SITE FOR THE P1-P3 GROUP OF A CLASS OF POTENT HIV-1 PROTEASE INHIBITORS AS A RESULT OF CONCERTED STRUCTURAL CHANGE IN 80'S LOOP. | | Descriptor: | (2S)-N-[(2S,3R)-4-[(2S,3S,4aS,8aS)-3-(tert-butylcarbamoyl)-3,4,4a,5,6,7,8,8a-octahydro-1H-isoquinolin-2-yl]-3-hydroxy-1 -phenyl-butan-2-yl]-2-(quinolin-2-ylcarbonylamino)butanediamide, PROTEIN (PROTEASE) | | Authors: | Munshi, S. | | Deposit date: | 1999-12-28 | | Release date: | 2000-12-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | An alternate binding site for the P1-P3 group of a class of potent HIV-1 protease inhibitors as a result of concerted structural change in the 80s loop of the protease.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
1C7H
 
 | | CRYSTAL STRUCTURE OF A MUTANT R75A IN KETOSTEROID ISOMERASE FROM PSEDOMONAS PUTIDA BIOTYPE B | | Descriptor: | DELTA-5-3-KETOSTEROID ISOMERASE | | Authors: | Nam, G.H, Kim, D.H, Jang, D.S, Choi, G, Ha, N.C, Oh, B.H, Choi, K.Y. | | Deposit date: | 2000-02-19 | | Release date: | 2000-04-24 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Roles of active site aromatic residues in catalysis by ketosteroid isomerase from Pseudomonas putida biotype B.
Biochemistry, 38, 1999
|
|
1C9W
 
 | | CHO REDUCTASE WITH NADP+ | | Descriptor: | CHO REDUCTASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Ye, Q, Li, X, Hyndman, D, Flynn, T.G, Jia, Z. | | Deposit date: | 1999-08-03 | | Release date: | 2000-01-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of CHO reductase, a member of the aldo-keto reductase superfamily.
Proteins, 38, 2000
|
|
1CAG
 
 | | CRYSTAL AND MOLECULAR STRUCTURE OF A COLLAGEN-LIKE PEPTIDE AT 1.9 ANGSTROM RESOLUTION | | Descriptor: | ACETIC ACID, COLLAGEN-LIKE PEPTIDE | | Authors: | Bella, J, Eaton, M, Brodsky, B, Berman, H.M. | | Deposit date: | 1994-03-29 | | Release date: | 1994-11-01 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal and molecular structure of a collagen-like peptide at 1.9 A resolution.
Science, 266, 1994
|
|
4B7E
 
 | | FACTOR INHIBITING HIF-1 ALPHA IN COMPLEX WITH CONSENSUS ANKYRIN REPEAT DOMAIN-LEU PEPTIDE (20-MER) | | Descriptor: | CONSENSUS ANKYRIN REPEAT DOMAIN-LEU, GLYCEROL, HYPOXIA-INDUCIBLE FACTOR 1-ALPHA INHIBITOR, ... | | Authors: | Chowdhury, R, Ge, W, Schofield, C.J. | | Deposit date: | 2012-08-17 | | Release date: | 2013-01-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Substrate selectivity analyses of factor inhibiting hypoxia-inducible factor.
Angew. Chem. Int. Ed. Engl., 52, 2013
|
|
1C6Y
 
 | |
1CC1
 
 | | CRYSTAL STRUCTURE OF A REDUCED, ACTIVE FORM OF THE NI-FE-SE HYDROGENASE FROM DESULFOMICROBIUM BACULATUM | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, FE (II) ION, HYDROGENASE (LARGE SUBUNIT), ... | | Authors: | Garcin, E, Vernede, X, Hatchikian, E.C, Volbeda, A, Frey, M, Fontecilla-Camps, J.C. | | Deposit date: | 1999-03-03 | | Release date: | 1999-06-01 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The crystal structure of a reduced [NiFeSe] hydrogenase provides an image of the activated catalytic center
Structure Fold.Des., 7, 1999
|
|
1CD3
 
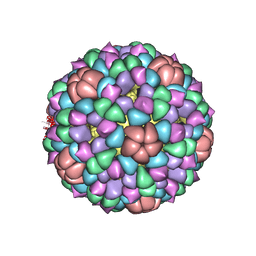 | | PROCAPSID OF BACTERIOPHAGE PHIX174 | | Descriptor: | PROTEIN (CAPSID PROTEIN GPF), PROTEIN (SCAFFOLDING PROTEIN GPB), PROTEIN (SCAFFOLDING PROTEIN GPD), ... | | Authors: | Rossmann, M.G, Dokland, T. | | Deposit date: | 1999-03-05 | | Release date: | 1999-04-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | The role of scaffolding proteins in the assembly of the small, single-stranded DNA virus phiX174.
J.Mol.Biol., 288, 1999
|
|
4B7K
 
 | | FACTOR INHIBITING HIF-1 ALPHA IN COMPLEX WITH CONSENSUS ANKYRIN REPEAT DOMAIN-SER PEPTIDE (20-MER) | | Descriptor: | CONSENSUS ANKYRIN REPEAT DOMAIN-SER, HYPOXIA-INDUCIBLE FACTOR 1-ALPHA INHIBITOR, N-OXALYLGLYCINE, ... | | Authors: | Chowdhury, R, Ge, W, Schofield, C.J. | | Deposit date: | 2012-08-20 | | Release date: | 2013-01-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Substrate selectivity analyses of factor inhibiting hypoxia-inducible factor.
Angew. Chem. Int. Ed. Engl., 52, 2013
|
|
4BH8
 
 | |
4BIO
 
 | |
4ARZ
 
 | | The crystal structure of Gtr1p-Gtr2p complexed with GTP-GDP | | Descriptor: | GTP-BINDING PROTEIN GTR1, GTP-BINDING PROTEIN GTR2, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Jeong, J.H, Kim, Y.G. | | Deposit date: | 2012-04-27 | | Release date: | 2012-07-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal Structure of the Gtr1Pgtp-Gtr2Pgdp Complex Reveals Large Structural Rearrangements Triggered by GTP-to-Gdp Conversion
J.Biol.Chem., 287, 2012
|
|
4B24
 
 | | Unprecedented sculpting of DNA at abasic sites by DNA glycosylase homolog Mag2 | | Descriptor: | 5'-D(*CP*GP*AP*TP*AP*GP*GP*TP*AP*GP)-3', 5'-D(*GP*CP*TP*AP*CP*3DRP*TP*AP*TP*CP*GP)-3', PROBABLE DNA-3-METHYLADENINE GLYCOSYLASE 2 | | Authors: | Dalhus, B, Nilsen, L, Korvald, H, Huffman, J, Forstrom, R.J, McMurray, C.T, Alseth, I, Tainer, J.A, Bjoras, M. | | Deposit date: | 2012-07-12 | | Release date: | 2013-01-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Sculpting of DNA at Abasic Sites by DNA Glycosylase Homolog Mag2.
Structure, 21, 2013
|
|
4B20
 
 | | Structural basis of DNA loop recognition by Endonuclease V | | Descriptor: | 5'-D(*AP*TP*CP*TP*TP*GP*TP*CP*GP*CP)-3', 5'-D(*GP*CP*GP*AP*CP*AP*GP)-3', ENDONUCLEASE V, ... | | Authors: | Rosnes, I, Rowe, A.D, Forstrom, R.J, Alseth, I, Bjoras, M, Dalhus, B. | | Deposit date: | 2012-07-12 | | Release date: | 2013-04-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural Basis of DNA Loop Recognition by Endonuclease V.
Structure, 21, 2013
|
|
4B23
 
 | | Unprecedented sculpting of DNA at abasic sites by DNA glycosylase homolog Mag2 | | Descriptor: | 5'-D(*CP*GP*AP*TP*TP*GP*GP*TP*AP*GP*CP)-3', 5'-D(*GP*CP*TP*AP*CP*(3DR)P*AP*AP*TP*CP*GP)-3', MAG2, ... | | Authors: | Dalhus, B, Nilsen, L, Korvald, H, Huffman, J, Forstrom, R.J, McMurray, C.T, Alseth, I, Tainer, J.A, Bjoras, M. | | Deposit date: | 2012-07-12 | | Release date: | 2013-01-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Sculpting of DNA at Abasic Sites by DNA Glycosylase Homolog Mag2.
Structure, 21, 2013
|
|
4ATW
 
 | | The crystal structure of Arabinofuranosidase | | Descriptor: | ALPHA-L-ARABINOFURANOSIDASE DOMAIN PROTEIN | | Authors: | Dumbrepatil, A, Song, H.-N, Jung, T.-Y, Kim, T.-J, Woo, E.-J. | | Deposit date: | 2012-05-10 | | Release date: | 2012-05-23 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Analysis of Alpha-L-Arabinofuranosidase from Thermotoga Maritima Reveals Characteristics for Thermostability and Substrate Specificity.
J.Microbiol.Biotech., 22, 2012
|
|
4BU2
 
 | | 60S ribosomal protein L27A histidine hydroxylase (MINA53) in complex with Ni(II) and 2-oxoglutarate (2OG) | | Descriptor: | 1,2-ETHANEDIOL, 2-OXOGLUTARIC ACID, BIFUNCTIONAL LYSINE-SPECIFIC DEMETHYLASE AND HISTIDYL-HYDROXYLASE MINA, ... | | Authors: | Chowdhury, R, Clifton, I.J, McDonough, M.A, Ng, S.S, Pilka, E, Oppermann, U, Schofield, C.J. | | Deposit date: | 2013-06-19 | | Release date: | 2014-05-14 | | Last modified: | 2019-02-06 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Ribosomal oxygenases are structurally conserved from prokaryotes to humans.
Nature, 510, 2014
|
|
4CEL
 
 | | ACTIVE-SITE MUTANT D214N DETERMINED AT PH 6.0 WITH NO LIGAND BOUND IN THE ACTIVE SITE | | Descriptor: | 1,4-BETA-D-GLUCAN CELLOBIOHYDROLASE I, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION | | Authors: | Divne, C, Stahlberg, J, Jones, T.A. | | Deposit date: | 1996-08-24 | | Release date: | 1997-03-12 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Activity studies and crystal structures of catalytically deficient mutants of cellobiohydrolase I from Trichoderma reesei.
J.Mol.Biol., 264, 1996
|
|
4BM5
 
 | | Chloroplast inner membrane protein TIC110 | | Descriptor: | SIMILAR TO CHLOROPLAST INNER MEMBRANE PROTEIN TIC110 | | Authors: | Tsai, J.-Y, Chu, C.-C, Yeh, Y.-H, Chen, L.-J, Li, H.-m, Hsiao, C.-D. | | Deposit date: | 2013-05-06 | | Release date: | 2013-06-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | Structural Characterizations of Chloroplast Translocon Protein Tic110.
Plant J., 75, 2013
|
|
