7DUB
 
 | |
1OYG
 
 | |
3UWS
 
 | |
7DZ0
 
 | |
1MUC
 
 | | STRUCTURE OF MUCONATE LACTONIZING ENZYME AT 1.85 ANGSTROMS RESOLUTION | | Descriptor: | MANGANESE (II) ION, MUCONATE LACTONIZING ENZYME | | Authors: | Helin, S, Kahn, P.C, Guha, B.H.L, Mallows, D.J, Goldman, A. | | Deposit date: | 1995-09-20 | | Release date: | 1996-07-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The refined X-ray structure of muconate lactonizing enzyme from Pseudomonas putida PRS2000 at 1.85 A resolution.
J.Mol.Biol., 254, 1995
|
|
2WM3
 
 | | Crystal structure of NmrA-like family domain containing protein 1 in complex with niflumic acid | | Descriptor: | 2-{[3-(TRIFLUOROMETHYL)PHENYL]AMINO}NICOTINIC ACID, CHLORIDE ION, GLYCEROL, ... | | Authors: | Bhatia, C, Yue, W.W, Niesen, F, Pilka, E, Ugochukwu, E, Savitsky, P, Hozjan, V, Roos, A.K, Filippakopoulos, P, von Delft, F, Heightman, T, Arrowsmith, C, Weigelt, J, Edwards, A, Bountra, C, Oppermann, U. | | Deposit date: | 2009-06-29 | | Release date: | 2009-08-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of Nmra-Like Family Domain Containing Protein 1 in Complex with Niflumic Acid
To be Published
|
|
4OQ8
 
 | | Satellite Tobacco Mosaic Virus Refined to 1.4 A Resolution using icosahedral constraints | | Descriptor: | Coat protein, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Larson, S.B, Day, J.S, McPherson, A. | | Deposit date: | 2014-02-07 | | Release date: | 2014-09-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Satellite tobacco mosaic virus refined to 1.4 angstrom resolution.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3BUH
 
 | | BACE-1 complexed with compound 4 | | Descriptor: | 4-(2-aminoethyl)-2-cyclohexylphenol, beta-secretase 1 | | Authors: | Kuglstatter, A, Hennig, M. | | Deposit date: | 2008-01-02 | | Release date: | 2008-03-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Tyramine fragment binding to BACE-1
Bioorg.Med.Chem.Lett., 18, 2008
|
|
2X58
 
 | | The crystal structure of MFE1 liganded with CoA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, COENZYME A, GLYCEROL, ... | | Authors: | Kasaragod, P, Venkatesan, R, Kiema, T.R, Hiltunen, J.K, Wierenga, R.K. | | Deposit date: | 2010-02-05 | | Release date: | 2010-05-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Crystal Structure of Liganded Rat Peroxisomal Multifunctional Enzyme Type 1: A Flexible Molecule with Two Interconnected Active Sites
J.Biol.Chem., 285, 2010
|
|
3C2X
 
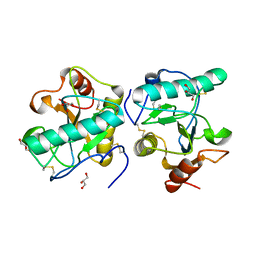 | | Crystal structure of peptidoglycan recognition protein at 1.8A resolution | | Descriptor: | GLYCEROL, L(+)-TARTARIC ACID, Peptidoglycan recognition protein, ... | | Authors: | Sharma, P, Singh, N, Sinha, M, Sharma, S, Perbandt, M, Betzel, C, Kaur, P, Srinivasan, A, Singh, T.P. | | Deposit date: | 2008-01-26 | | Release date: | 2008-03-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Crystal structure of the peptidoglycan recognition protein at 1.8 A resolution reveals dual strategy to combat infection through two independent functional homodimers
J.Mol.Biol., 378, 2008
|
|
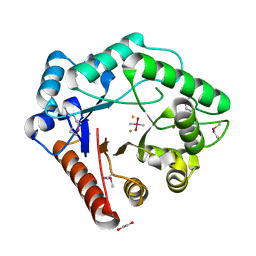
3BWW
 
 | |
1U8O
 
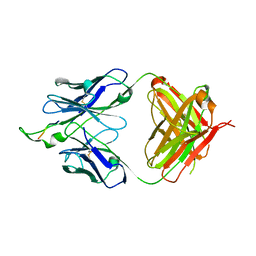 | | Crystal structure of the HIV-1 Cross Neutralizing Monoclonal Antibody 2F5 in complex with gp41 Peptide ELDKHAS | | Descriptor: | ANTIBODY 2F5 (HEAVY CHAIN), ANTIBODY 2F5 (LIGHT CHAIN), GP41 PEPTIDE | | Authors: | Bryson, S, Julien, J.-P, Hynes, R.C, Pai, E.F. | | Deposit date: | 2004-08-06 | | Release date: | 2004-10-05 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | Crystallographic definition of the epitope promiscuity of the broadly neutralizing anti-human immunodeficiency virus type 1 antibody 2F5: vaccine design implications
J.Virol., 83, 2009
|
|
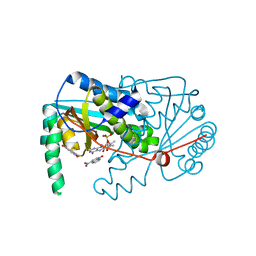
4BNB
 
 | |
3U16
 
 | | Structure of BasE N-terminal domain from Acinetobacter baumannii bound to 6-(p-benzyloxy)phenyl-1-(pyridin-4-ylmethyl)-1H-pyrazolo[3,4-b]pyridine-4-carboxylic acid. | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 6-[4-(benzyloxy)phenyl]-1-(pyridin-4-ylmethyl)-1H-pyrazolo[3,4-b]pyridine-4-carboxylic acid, ... | | Authors: | Gulick, A.M, Drake, E.J, Aldrich, C.C, Neres, J. | | Deposit date: | 2011-09-29 | | Release date: | 2012-10-03 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Non-nucleoside inhibitors of BasE, an adenylating enzyme in the siderophore biosynthetic pathway of the opportunistic pathogen Acinetobacter baumannii.
J.Med.Chem., 56, 2013
|
|
1ZB6
 
 | | Co-Crystal Structure of ORF2 an Aromatic Prenyl Transferase from Streptomyces sp. strain cl190 complexed with GSPP and 1,6-dihydroxynaphtalene | | Descriptor: | 1,6-DIHYDROXY NAPHTHALENE, Aromatic prenyltransferase, GERANYL S-THIOLODIPHOSPHATE, ... | | Authors: | Kuzuyama, T, Noel, J.P, Richard, S.B. | | Deposit date: | 2005-04-07 | | Release date: | 2005-06-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis for the promiscuous biosynthetic prenylation of aromatic natural products.
Nature, 435, 2005
|
|
2JEP
 
 | | Native family 5 xyloglucanase from Paenibacillus pabuli | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, XYLOGLUCANASE | | Authors: | Gloster, T.M, Ibatullin, F.M, Macauley, K, Eklof, J.M, Roberts, S, Turkenburg, J.P, Bjornvad, M.E, Jorgensen, P.L, Danielsen, S, Johansen, K, Borchert, T.V, Wilson, K.S, Brumer, H, Davies, G.J. | | Deposit date: | 2007-01-18 | | Release date: | 2007-03-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Characterization and Three-Dimensional Structures of Two Distinct Bacterial Xyloglucanases from Families Gh5 and Gh12.
J.Biol.Chem., 282, 2007
|
|
1EK6
 
 | | STRUCTURE OF HUMAN UDP-GALACTOSE 4-EPIMERASE COMPLEXED WITH NADH AND UDP-GLUCOSE | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, MAGNESIUM ION, TETRAMETHYLAMMONIUM ION, ... | | Authors: | Thoden, J.B, Wohlers, T.M, Fridovich-Keil, J.L, Holden, H.M. | | Deposit date: | 2000-03-06 | | Release date: | 2000-05-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystallographic evidence for Tyr 157 functioning as the active site base in human UDP-galactose 4-epimerase.
Biochemistry, 39, 2000
|
|
4YRI
 
 | |
6JSG
 
 | | Crystal Structure of BACE1 in complex with N-{3-[(4S)-2-amino-4-methyl-5,6-dihydro-4H-1,3-thiazin-4-yl]-4-fluorophenyl}-5-chloropyridine-2-carboxamide | | Descriptor: | Beta-secretase 1, GLYCEROL, IODIDE ION, ... | | Authors: | Fujimoto, K, Matsuoka, E, Asada, N, Tadano, G, Yamamoto, T, Nakahara, K, Fuchino, K, Ito, H, Kanegawa, N, Moechars, D, Gijsen, H.J.M, Kusakabe, K.I. | | Deposit date: | 2019-04-08 | | Release date: | 2019-08-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-Based Design of Selective beta-Site Amyloid Precursor Protein Cleaving Enzyme 1 (BACE1) Inhibitors: Targeting the Flap to Gain Selectivity over BACE2.
J.Med.Chem., 62, 2019
|
|
6S8U
 
 | | Structure of the PfEMP1 IT4var13 DBLbeta domain bound to ICAM-1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Erythrocyte membrane protein 1, ... | | Authors: | Lennartz, F, Higgins, M.K. | | Deposit date: | 2019-07-10 | | Release date: | 2019-09-25 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.67 Å) | | Cite: | Structural insights into diverse modes of ICAM-1 binding byPlasmodium falciparum-infected erythrocytes.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
1K30
 
 | | Crystal Structure Analysis of Squash (Cucurbita moschata) glycerol-3-phosphate (1)-acyltransferase | | Descriptor: | glycerol-3-phosphate acyltransferase | | Authors: | Turnbull, A.P, Rafferty, J.B, Sedelnikova, S.E, Slabas, A.R, Schierer, T.P, Kroon, J.T, Simon, J.W, Fawcett, T, Nishida, I, Murata, N, Rice, D.W. | | Deposit date: | 2001-10-01 | | Release date: | 2001-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Analysis of the structure, substrate specificity, and mechanism of squash glycerol-3-phosphate (1)-acyltransferase.
Structure, 9, 2001
|
|
4OGY
 
 | | Crystal structure of Fab DX-2930 in complex with human plasma kallikrein at 2.1 Angstrom resolution | | Descriptor: | 1,2-ETHANEDIOL, DX-2930 HEAVY CHAIN, DX-2930 LIGHT CHAIN, ... | | Authors: | Edwards, T.E, Clifton, M.C, Abendroth, J, Nixon, A, Ladner, R. | | Deposit date: | 2014-01-16 | | Release date: | 2014-07-09 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Inhibition of plasma kallikrein by a highly specific active site blocking antibody.
J.Biol.Chem., 289, 2014
|
|
4YRP
 
 | |
4LSV
 
 | | Crystal structure of broadly and potently neutralizing antibody 3BNC117 in complex with HIV-1 clade C C1086 gp120 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HEAVY CHAIN OF ANTIBODY 3BNC117, IMIDAZOLE, ... | | Authors: | Zhou, T, Moquin, S, Kwong, P.D. | | Deposit date: | 2013-07-23 | | Release date: | 2013-08-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Multidonor Analysis Reveals Structural Elements, Genetic Determinants, and Maturation Pathway for HIV-1 Neutralization by VRC01-Class Antibodies.
Immunity, 39, 2013
|
|
1JL0
 
 | | Structure of a Human S-Adenosylmethionine Decarboxylase Self-processing Ester Intermediate and Mechanism of Putrescine Stimulation of Processing as Revealed by the H243A Mutant | | Descriptor: | 1,4-DIAMINOBUTANE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, S-ADENOSYLMETHIONINE DECARBOXYLASE PROENZYME | | Authors: | Ekstrom, J.L, Tolbert, W.D, Xiong, H, Pegg, A.E, Ealick, S.E. | | Deposit date: | 2001-07-13 | | Release date: | 2001-08-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of a human S-adenosylmethionine decarboxylase self-processing ester intermediate and mechanism of putrescine stimulation of processing as revealed by the H243A mutant.
Biochemistry, 40, 2001
|
|
