6MK5
 
 | |
6MZT
 
 | | Solution structure of alpha-KTx-6.21 (UroTx) from Urodacus yaschenkoi | | Descriptor: | Potassium channel toxin alpha-KTx 6.21 | | Authors: | Chin, Y.K.-Y, Luna-Ramirez, K, Anangi, R, King, G.F. | | Deposit date: | 2018-11-05 | | Release date: | 2020-03-11 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Structural basis of the potency and selectivity of Urotoxin, a potent Kv1 blocker from scorpion venom.
Biochem. Pharmacol., 174, 2020
|
|
8IO9
 
 | |
8RPZ
 
 | | Escherichia coli 50S subunit in complex with the antimicrobial peptide Api88 - conformation I | | Descriptor: | 23S ribosomal RNA, 5S ribosomal RNA, Apidaecins type 88, ... | | Authors: | Lauer, S, Nikolay, R, Spahn, C. | | Deposit date: | 2024-01-17 | | Release date: | 2024-05-22 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.44 Å) | | Cite: | Multimodal binding and inhibition of bacterial ribosomes by the antimicrobial peptides Api137 and Api88.
Nat Commun, 15, 2024
|
|
8RPY
 
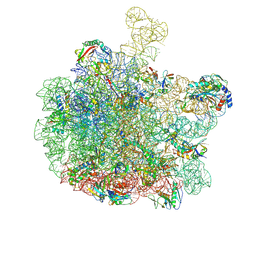 | | Escherichia coli 50S subunit in complex with the antimicrobial peptide Api137 | | Descriptor: | 23S ribosomal RNA, 5S ribosomal RNA, Apidaecins type 137, ... | | Authors: | Lauer, S, Nikolay, R, Spahn, C. | | Deposit date: | 2024-01-17 | | Release date: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.64 Å) | | Cite: | Multimodal binding and inhibition of bacterial ribosomes by the antimicrobial peptides Api137 and Api88.
Nat Commun, 15, 2024
|
|
8RQ0
 
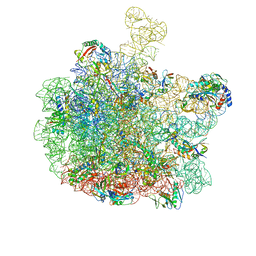 | | Escherichia coli 50S subunit in complex with the antimicrobial peptide Api88 - conformation II | | Descriptor: | 23S ribosomal RNA, 5S ribosomal RNA, Apidaecins type 88, ... | | Authors: | Lauer, S, Nikolay, R, Spahn, C. | | Deposit date: | 2024-01-17 | | Release date: | 2024-05-22 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.44 Å) | | Cite: | Multimodal binding and inhibition of bacterial ribosomes by the antimicrobial peptides Api137 and Api88.
Nat Commun, 15, 2024
|
|
5OK9
 
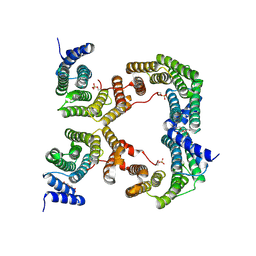 | | CH1 chimera of human 14-3-3 sigma with the HSPB6 phosphopeptide in a conformation with swapped phosphopeptides | | Descriptor: | 14-3-3 protein sigma,Heat shock protein beta-6, DI(HYDROXYETHYL)ETHER | | Authors: | Sluchanko, N.N, Tugaeva, K.V, Greive, S.J, Antson, A.A. | | Deposit date: | 2017-07-25 | | Release date: | 2017-10-11 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Chimeric 14-3-3 proteins for unraveling interactions with intrinsically disordered partners.
Sci Rep, 7, 2017
|
|
6JIC
 
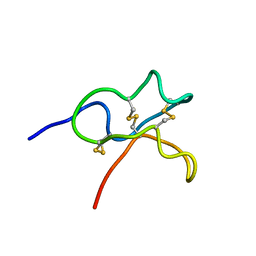 | | Identification and Characterization of a carboxypeptidase inhibitor from Lycium barbarum | | Descriptor: | WCI | | Authors: | Tan, W.L, Wong, K.H, Huang, J.Y, Tay, S.V, Wang, S.J. | | Deposit date: | 2019-02-20 | | Release date: | 2020-02-26 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Identification and characterization of a wolfberry carboxypeptidase inhibitor from Lycium barbarum.
Food Chem, 351, 2021
|
|
8G5H
 
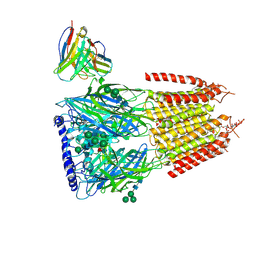 | | Native GABA-A receptor from the mouse brain, ortho-alpha1-alpha3-beta2-gamma2 subtype, in complex with GABA, Zolpidem, and endogenous neurosteroid | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GAMMA-AMINO-BUTANOIC ACID, Gamma-aminobutyric acid receptor subunit alpha-1, ... | | Authors: | Sun, C, Gouaux, E. | | Deposit date: | 2023-02-13 | | Release date: | 2023-09-20 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Cryo-EM structures reveal native GABA A receptor assemblies and pharmacology.
Nature, 622, 2023
|
|
1FSS
 
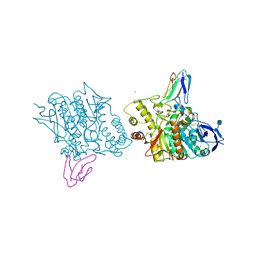 | | ACETYLCHOLINESTERASE (E.C. 3.1.1.7) COMPLEXED WITH FASCICULIN-II | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYLCHOLINESTERASE, FASCICULIN II, ... | | Authors: | Harel, M, Kleywegt, G.J, Silman, I, Sussman, J.L. | | Deposit date: | 1995-10-25 | | Release date: | 1996-03-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of an acetylcholinesterase-fasciculin complex: interaction of a three-fingered toxin from snake venom with its target.
Structure, 3, 1995
|
|
8G2U
 
 | | Time-resolved cryo-EM study of the 70S recycling by the HflX:control-apo-70S at 900ms | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Bhattacharjee, S, Brown, P.Z, Frank, J. | | Deposit date: | 2023-02-06 | | Release date: | 2023-12-06 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Time resolution in cryo-EM using a PDMS-based microfluidic chip assembly and its application to the study of HflX-mediated ribosome recycling.
Cell, 187, 2024
|
|
8G34
 
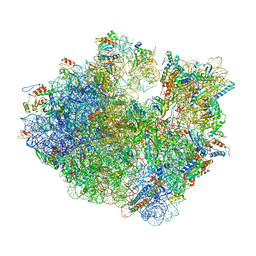 | | Time-resolved cryo-EM study of the 70S recycling by the HflX:1st intermediate | | Descriptor: | 16S, 23S, 30S ribosomal protein S10, ... | | Authors: | Bhattacharjee, S, Brown, P.Z, Frank, J. | | Deposit date: | 2023-02-06 | | Release date: | 2023-12-06 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Time resolution in cryo-EM using a PDMS-based microfluidic chip assembly and its application to the study of HflX-mediated ribosome recycling.
Cell, 187, 2024
|
|
8G31
 
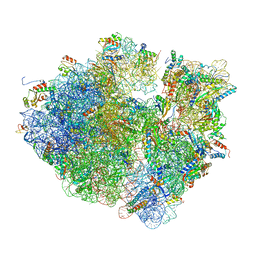 | | Time-resolved cryo-EM study of the 70S recycling by the HflX:2nd Intermediate | | Descriptor: | 16S, 23S, 30S ribosomal protein S10, ... | | Authors: | Bhattacharjee, S, Brown, P.Z, Frank, J. | | Deposit date: | 2023-02-06 | | Release date: | 2023-12-06 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Time resolution in cryo-EM using a PDMS-based microfluidic chip assembly and its application to the study of HflX-mediated ribosome recycling.
Cell, 187, 2024
|
|
5OKF
 
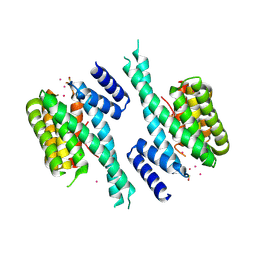 | | CH1 chimera of human 14-3-3 sigma with the HSPB6 phosphopeptide in a conformation with self-bound phosphopeptides | | Descriptor: | 14-3-3 protein sigma,Heat shock protein beta-6, CADMIUM ION | | Authors: | Sluchanko, N.N, Tugaeva, K.V, Greive, S.J, Antson, A.A. | | Deposit date: | 2017-07-25 | | Release date: | 2017-10-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Chimeric 14-3-3 proteins for unraveling interactions with intrinsically disordered partners.
Sci Rep, 7, 2017
|
|
4MUW
 
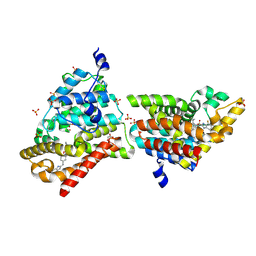 | | Crystal Structure of PDE10A with Novel Keto-Benzimidazole Inhibitor | | Descriptor: | 2-{4-[(6,7-difluoro-1H-benzimidazol-2-yl)amino]phenoxy}-N-methyl-3,4'-bipyridin-2'-amine, GLYCEROL, SULFATE ION, ... | | Authors: | Chmait, S, Jordan, S. | | Deposit date: | 2013-09-23 | | Release date: | 2013-10-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.639 Å) | | Cite: | Design, Optimization, and Biological Evaluation of Novel Keto-Benzimidazoles as Potent and Selective Inhibitors of Phosphodiesterase 10A (PDE10A).
J.Med.Chem., 56, 2013
|
|
6GNZ
 
 | |
6CEI
 
 | |
6CFB
 
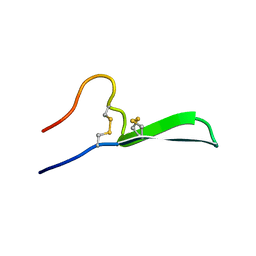 | | Isolation, Characterization, and Synthesis of the Barrettides: Disulfide-Containing Peptides from the Marine Sponge Geodia barretti | | Descriptor: | barrettide A | | Authors: | Rosengren, K.J, Carstens, B.B, Clark, R.J, Goransson, U. | | Deposit date: | 2018-02-14 | | Release date: | 2018-03-21 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Isolation, Characterization, and Synthesis of the Barrettides: Disulfide-Containing Peptides from the Marine Sponge Geodia barretti.
J. Nat. Prod., 78, 2015
|
|
6O3S
 
 | | NMR solution structure of Luffin P1 | | Descriptor: | Ribosome-inactivating protein luffin P1 | | Authors: | Rosengren, K.J, Payne, C. | | Deposit date: | 2019-02-27 | | Release date: | 2019-04-24 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | An Ancient Peptide Family Buried within Vicilin Precursors.
Acs Chem.Biol., 14, 2019
|
|
8TWB
 
 | | Cryo-EM structure of S. cerevisiae Ctf18-RFC-PCNA-DNA complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Chromosome transmission fidelity protein 18, MAGNESIUM ION, ... | | Authors: | Yuan, Z, Georgescu, R, O'Donnell, M, Li, H. | | Deposit date: | 2023-08-20 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Mechanism of PCNA loading by Ctf18-RFC for leading-strand DNA synthesis.
Science, 385, 2024
|
|
6O3Q
 
 | |
8TW7
 
 | | Cryo-EM structure of S. cerevisiae Ctf18-RFC-PCNA complex in Apo state conformation I | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Chromosome transmission fidelity protein 18, MAGNESIUM ION, ... | | Authors: | Yuan, Z, Georgescu, R, O'Donnell, M, Li, H. | | Deposit date: | 2023-08-20 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Mechanism of PCNA loading by Ctf18-RFC for leading-strand DNA synthesis.
Science, 385, 2024
|
|
8TWA
 
 | | Cryo-EM structure of S. cerevisiae Ctf18-RFC-PCNA-PolE-DNA complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Chromosome transmission fidelity protein 18, Chromosome transmission fidelity protein 8, ... | | Authors: | Yuan, Z, Georgescu, R, O'Donnell, M, Li, H. | | Deposit date: | 2023-08-20 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Mechanism of PCNA loading by Ctf18-RFC for leading-strand DNA synthesis.
Science, 385, 2024
|
|
8TW8
 
 | | Cryo-EM structure of S. cerevisiae Ctf18-RFC-PCNA complex in Apo state conformation I | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Chromosome transmission fidelity protein 18, MAGNESIUM ION, ... | | Authors: | Yuan, Z, Georgescu, R, O'Donnell, M, Li, H. | | Deposit date: | 2023-08-20 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Mechanism of PCNA loading by Ctf18-RFC for leading-strand DNA synthesis.
Science, 385, 2024
|
|
7N7E
 
 | |
