3JSB
 
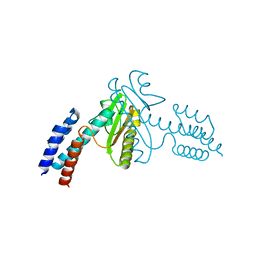 | | Crystal structure of the N-terminal domain of the Lymphocytic Choriomeningitis Virus L protein | | Descriptor: | RNA-directed RNA polymerase | | Authors: | Morin, B, Jamal, S, Ferron, F.P, Coutard, B, Bricogne, G, Canard, B, Vonrhein, C, Marseilles Structural Genomics Program @ AFMB (MSGP) | | Deposit date: | 2009-09-10 | | Release date: | 2010-09-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | The N-terminal domain of the arenavirus L protein is an RNA endonuclease essential in mRNA transcription
Plos Pathog., 6, 2010
|
|
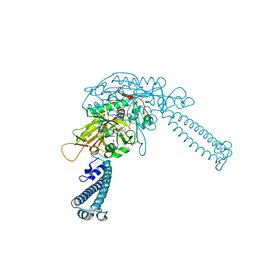
4L87
 
 | |
7QDY
 
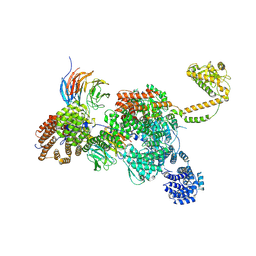 | | RNA-bound human SKI complex | | Descriptor: | Helicase SKI2W, RNA (5'-R(P*UP*UP*UP*UP*UP*U)-3'), Tetratricopeptide repeat protein 37, ... | | Authors: | Koegel, A, Keidel, A, Bonneau, F, Schaefer, I.B, Conti, E. | | Deposit date: | 2021-12-01 | | Release date: | 2022-02-09 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | The human SKI complex regulates channeling of ribosome-bound RNA to the exosome via an intrinsic gatekeeping mechanism.
Mol.Cell, 82, 2022
|
|
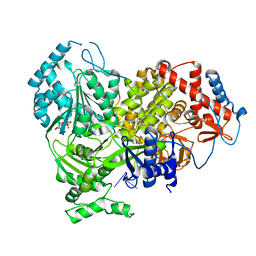
8B9I
 
 | |
8B9G
 
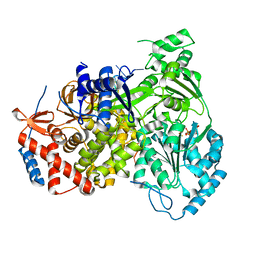 | | Cryo-EM structure of MLE in complex with ADP:AlF4 and U10 RNA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Dosage compensation regulator, RNA (5'-R(P*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*U)-3'), ... | | Authors: | Jagtap, P.K.A, Hennig, J. | | Deposit date: | 2022-10-06 | | Release date: | 2023-10-18 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | Structural basis of RNA-induced autoregulation of the DExH-type RNA helicase maleless.
Mol.Cell, 83, 2023
|
|
4PTF
 
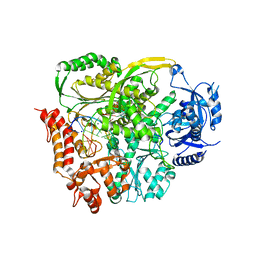 | | Ternary crystal structure of yeast DNA polymerase epsilon with template G | | Descriptor: | 1,2-ETHANEDIOL, 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, 5'-D(*AP*TP*CP*CP*TP*CP*CP*CP*CP*TP*AP*(DOC))-3', ... | | Authors: | Jain, R, Rajashankar, K.R, Buku, A, Johnson, R.E, Prakash, L, Prakash, S, Aggarwal, A.K. | | Deposit date: | 2014-03-10 | | Release date: | 2014-04-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.809 Å) | | Cite: | Crystal Structure of Yeast DNA Polymerase epsilon Catalytic Domain.
Plos One, 9, 2014
|
|
2M23
 
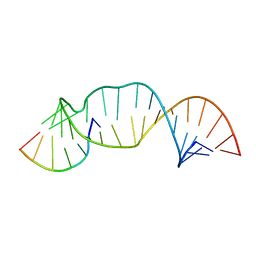 | |
2M24
 
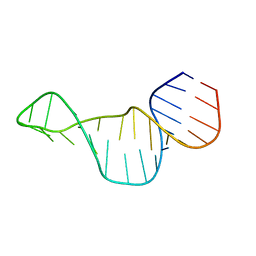 | |
9EAL
 
 | |
8P4F
 
 | | Structural insights into human co-transcriptional capping - structure 6 | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-TRIPHOSPHATE, Cap-specific mRNA (nucleoside-2'-O-)-methyltransferase 1, DNA (38-MER), ... | | Authors: | Garg, G, Dienemann, C, Farnung, L, Schwarz, J, Linden, A, Urlaub, H, Cramer, P. | | Deposit date: | 2023-05-20 | | Release date: | 2023-07-19 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural insights into human co-transcriptional capping.
Mol.Cell, 83, 2023
|
|
8P4E
 
 | | Structural insights into human co-transcriptional capping - structure 5 | | Descriptor: | Cap-specific mRNA (nucleoside-2'-O-)-methyltransferase 1, DNA (26-MER), DNA (35-MER), ... | | Authors: | Garg, G, Dienemann, C, Farnung, L, Schwarz, J, Linden, A, Urlaub, H, Cramer, P. | | Deposit date: | 2023-05-20 | | Release date: | 2023-07-19 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural insights into human co-transcriptional capping.
Mol.Cell, 83, 2023
|
|
8P4C
 
 | | Structural insights into human co-transcriptional capping - structure 3 | | Descriptor: | DNA (32-MER), DNA (41-MER), DNA-directed RNA polymerase II subunit E, ... | | Authors: | Garg, G, Dienemann, C, Farnung, L, Schwarz, J, Linden, A, Urlaub, H, Cramer, P. | | Deposit date: | 2023-05-20 | | Release date: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural insights into human co-transcriptional capping.
Mol.Cell, 83, 2023
|
|
8P4D
 
 | | Structural insights into human co-transcriptional capping - structure 4 | | Descriptor: | DNA (32-MER), DNA (41-MER), DNA-directed RNA polymerase II subunit E, ... | | Authors: | Garg, G, Dienemann, C, Farnung, L, Schwarz, J, Linden, A, Urlaub, H, Cramer, P. | | Deposit date: | 2023-05-20 | | Release date: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural insights into human co-transcriptional capping.
Mol.Cell, 83, 2023
|
|
8P4B
 
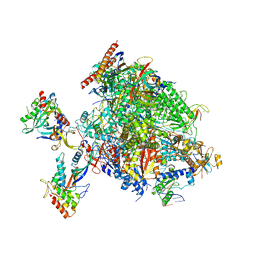 | | Structural insights into human co-transcriptional capping - structure 2 | | Descriptor: | DNA (35-MER), DNA (5'-D(P*AP*GP*CP*GP*GP*CP*GP*GP*AP*GP*CP*CP*AP*GP*CP*AP*GP*GP*GP*AP*GP*CP*TP*G)-3'), DNA-directed RNA polymerase II subunit E, ... | | Authors: | Garg, G, Dienemann, C, Farnung, L, Schwarz, J, Linden, A, Urlaub, H, Cramer, P. | | Deposit date: | 2023-05-20 | | Release date: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insights into human co-transcriptional capping.
Mol.Cell, 83, 2023
|
|
8P4A
 
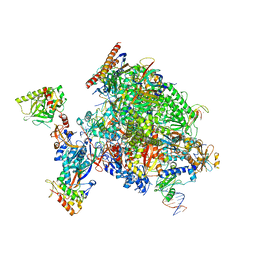 | | Structural insights into human co-transcriptional capping - structure 1 | | Descriptor: | DNA (29-MER), DNA (38-MER), DNA-directed RNA polymerase II subunit E, ... | | Authors: | Garg, G, Dienemann, C, Farnung, L, Schwarz, J, Linden, A, Urlaub, H, Cramer, P. | | Deposit date: | 2023-05-20 | | Release date: | 2023-08-16 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural insights into human co-transcriptional capping.
Mol.Cell, 83, 2023
|
|
6CCE
 
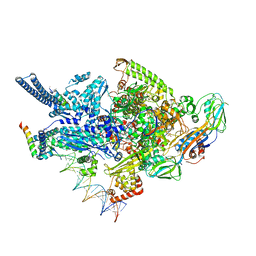 | | Crystal structure of a Mycobacterium smegmatis RNA polymerase transcription initiation complex with inhibitor Kanglemycin A | | Descriptor: | 1,2-ETHANEDIOL, DNA (57-MER), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Lilic, M, Darst, S.A, Campbell, E.A. | | Deposit date: | 2018-02-07 | | Release date: | 2018-08-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Rifamycin congeners kanglemycins are active against rifampicin-resistant bacteria via a distinct mechanism.
Nat Commun, 9, 2018
|
|
5D4E
 
 | |
6PB4
 
 | |
6PB5
 
 | |
7ABG
 
 | | Human pre-Bact-1 spliceosome | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE-5'-GUANOSINE, Cell division cycle 5-like protein, ... | | Authors: | Townsend, C, Kastner, B, Leelaram, M.N, Bertram, K, Stark, H, Luehrmann, R. | | Deposit date: | 2020-09-07 | | Release date: | 2020-12-23 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (7.8 Å) | | Cite: | Mechanism of protein-guided folding of the active site U2/U6 RNA during spliceosome activation.
Science, 370, 2020
|
|
8PH9
 
 | | E. coli RNA polymerase paused at ops site (non-complementary scaffold) | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Zuber, P.K, Said, N, Hilal, T, Loll, B, Wahl, M.C, Knauer, S.H. | | Deposit date: | 2023-06-19 | | Release date: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Concerted transformation of a hyper-paused transcription complex and its reinforcing protein.
Nat Commun, 15, 2024
|
|
8PDY
 
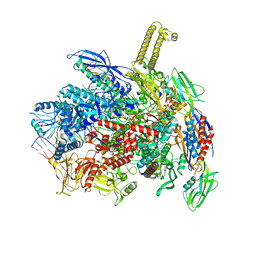 | | E. coli RNA polymerase paused at ops site | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Zuber, P.K, Said, N, Hilal, T, Loll, B, Wahl, M.C, Knauer, S.H. | | Deposit date: | 2023-06-13 | | Release date: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Concerted transformation of a hyper-paused transcription complex and its reinforcing protein.
Nat Commun, 15, 2024
|
|
9F37
 
 | |
8YFR
 
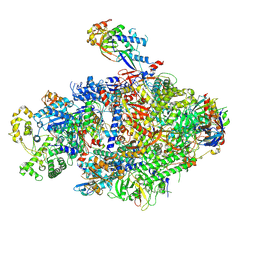 | | Cryo EM structure of Komagataella phaffii Rat1-Rai1 complex bound within the RNAPII cleft | | Descriptor: | 5'-3' exoribonuclease, DNA-directed RNA polymerase subunit, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Murayama, Y, Yanagisawa, T, Ehara, H, Sekine, S. | | Deposit date: | 2024-02-25 | | Release date: | 2024-09-25 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis of eukaryotic transcription termination by the Rat1 exonuclease complex.
Nat Commun, 15, 2024
|
|
8YFQ
 
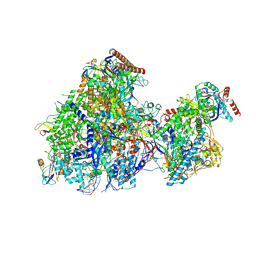 | | Cryo EM structure of Komagataella phaffii RNAPII-Rat1-Rai1 pre-termination complex | | Descriptor: | 5'-3' exoribonuclease, DNA (90-mer), DNA-directed RNA polymerase subunit, ... | | Authors: | Murayama, Y, Yanagisawa, T, Ehara, H, Sekine, S. | | Deposit date: | 2024-02-25 | | Release date: | 2024-09-25 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis of eukaryotic transcription termination by the Rat1 exonuclease complex.
Nat Commun, 15, 2024
|
|
