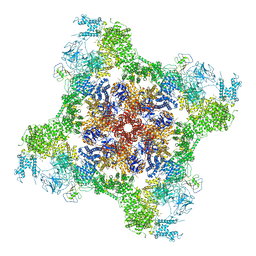
7U9Z
 
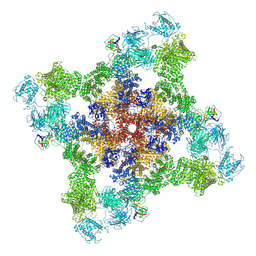 | |
7T64
 
 | | Rabbit RyR1 disease mutant Y523S in complex with FKBP12.6 embedded in lipidic nanodisc in the closed state | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 1, ZINC ION | | Authors: | Iyer, K.A, Hu, Y, Murayama, T, Samso, M. | | Deposit date: | 2021-12-13 | | Release date: | 2022-07-20 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Molecular mechanism of the severe MH/CCD mutation Y522S in skeletal ryanodine receptor (RyR1) by cryo-EM.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
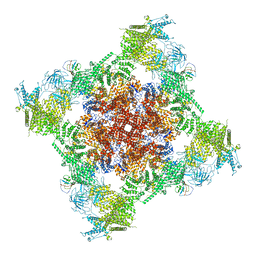
7T65
 
 | | Rabbit RyR1 disease mutant Y523S in complex with FKBP12.6 embedded in lipidic nanodisc in the open state | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, Peptidyl-prolyl cis-trans isomerase FKBP1B, ... | | Authors: | Iyer, K.A, Hu, Y, Murayama, T, Samso, M. | | Deposit date: | 2021-12-13 | | Release date: | 2022-07-20 | | Last modified: | 2022-08-03 | | Method: | ELECTRON MICROSCOPY (4.05 Å) | | Cite: | Molecular mechanism of the severe MH/CCD mutation Y522S in skeletal ryanodine receptor (RyR1) by cryo-EM.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7OW2
 
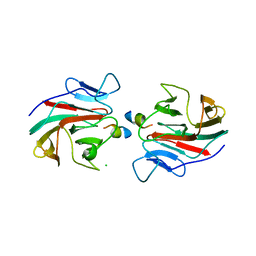 | | E3 RING ligase binding domain with peptide | | Descriptor: | CHLORIDE ION, E3 ubiquitin-protein ligase RNF187 peptide, E3 ubiquitin-protein ligase TRIM7, ... | | Authors: | James, L.C. | | Deposit date: | 2021-06-16 | | Release date: | 2022-07-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | E3 ligase targeting domain
To Be Published
|
|
8A8X
 
 | | TRIM7 PRYSPRY in complex with a MNV1-NS3 peptide HDDFGLQ | | Descriptor: | E3 ubiquitin-protein ligase TRIM7, MNV1-NS3 C term peptide | | Authors: | Luptak, J. | | Deposit date: | 2022-06-24 | | Release date: | 2022-07-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | TRIM7 Restricts Coxsackievirus and Norovirus Infection by Detecting the C-Terminal Glutamine Generated by 3C Protease Processing.
Viruses, 14, 2022
|
|
8A5M
 
 | | TRIM7 PRYSPRY in complex with a MNV1-NS6 peptide LEALEFQ | | Descriptor: | E3 ubiquitin-protein ligase TRIM7, MNV1-NS6 peptide LEALEFQ, PHOSPHATE ION | | Authors: | Luptak, J. | | Deposit date: | 2022-06-15 | | Release date: | 2022-07-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.918 Å) | | Cite: | TRIM7 Restricts Coxsackievirus and Norovirus Infection by Detecting the C-Terminal Glutamine Generated by 3C Protease Processing.
Viruses, 14, 2022
|
|
8A5L
 
 | | TRIM7 PRYSPRY in complex with a 2BC peptide TIEALFQ | | Descriptor: | 2BC peptide TIEALFQ, D(-)-TARTARIC ACID, E3 ubiquitin-protein ligase TRIM7 | | Authors: | Luptak, J. | | Deposit date: | 2022-06-15 | | Release date: | 2022-07-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.622 Å) | | Cite: | TRIM7 Restricts Coxsackievirus and Norovirus Infection by Detecting the C-Terminal Glutamine Generated by 3C Protease Processing.
Viruses, 14, 2022
|
|
7OVX
 
 | | E3 RING ligase binding domain | | Descriptor: | E3 ubiquitin-protein ligase TRIM7, Peptide G | | Authors: | James, L.C. | | Deposit date: | 2021-06-15 | | Release date: | 2022-06-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | E3 ligase targeting domain
To Be Published
|
|
7WUG
 
 | | GID subcomplex: Gid12 bound Substrate Receptor Scaffolding module | | Descriptor: | Glucose-induced degradation protein 8, HLJ1_G0042170.mRNA.1.CDS.1, Vacuolar import and degradation protein 24, ... | | Authors: | Qiao, S, Cheng, J.D, Schulman, B.A. | | Deposit date: | 2022-02-08 | | Release date: | 2022-06-08 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures of Gid12-bound GID E3 reveal steric blockade as a mechanism inhibiting substrate ubiquitylation.
Nat Commun, 13, 2022
|
|
7TZC
 
 | | A drug and ATP binding site in type 1 ryanodine receptor | | Descriptor: | (2S)-3-(octadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 4-[(7-methoxy-2,3-dihydro-1,4-benzothiazepin-4(5H)-yl)methyl]benzoic acid, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Melville, Z, Dridi, H, Yuan, Q, Reiken, S, Anetta, W, Liu, Y, Clarke, O.B, Marks, A.R. | | Deposit date: | 2022-02-15 | | Release date: | 2022-05-18 | | Last modified: | 2022-07-20 | | Method: | ELECTRON MICROSCOPY (2.45 Å) | | Cite: | A drug and ATP binding site in type 1 ryanodine receptor.
Structure, 30, 2022
|
|
7QRZ
 
 | |
7QS2
 
 | |
7QS5
 
 | |
7QS3
 
 | |
7QRY
 
 | |
7QS1
 
 | |
7QS0
 
 | |
7TDJ
 
 | | Rabbit RyR1 with AMP-PCP and high Ca2+ embedded in nanodisc in closed-inactivated conformation class 1(Dataset-A) | | Descriptor: | CALCIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, Ryanodine receptor 1,Ryanodine receptor 1,RyR1, ... | | Authors: | Nayak, A.R, Samso, M. | | Deposit date: | 2021-12-31 | | Release date: | 2022-03-09 | | Last modified: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Ca 2+ -inactivation of the mammalian ryanodine receptor type 1 in a lipidic environment revealed by cryo-EM.
Elife, 11, 2022
|
|
7TDG
 
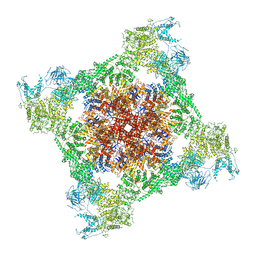 | |
7TDH
 
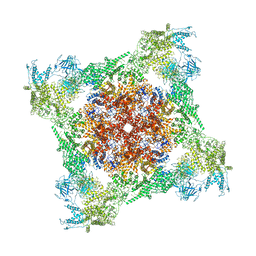 | | Rabbit RyR1 with AMP-PCP and high Ca2+ embedded in nanodisc in open conformation | | Descriptor: | CALCIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, Ryanodine receptor 1,Ryanodine receptor 1,RyR1, ... | | Authors: | Nayak, A.R, Samso, M. | | Deposit date: | 2021-12-31 | | Release date: | 2022-03-09 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Ca 2+ -inactivation of the mammalian ryanodine receptor type 1 in a lipidic environment revealed by cryo-EM.
Elife, 11, 2022
|
|
7TDI
 
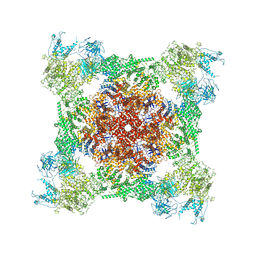 | | Rabbit RyR1 with AMP-PCP and high Ca2+ embedded in nanodisc in closed-inactivated conformation class 2 (Dataset-A) | | Descriptor: | CALCIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, Ryanodine receptor 1,Ryanodine receptor 1,RyR1, ... | | Authors: | Nayak, A.R, Samso, M. | | Deposit date: | 2021-12-31 | | Release date: | 2022-03-09 | | Last modified: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Ca 2+ -inactivation of the mammalian ryanodine receptor type 1 in a lipidic environment revealed by cryo-EM.
Elife, 11, 2022
|
|
7TDK
 
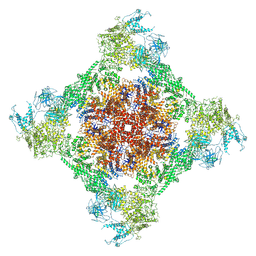 | | Rabbit RyR1 with AMP-PCP and high Ca2+ embedded in nanodisc in closed-inactivated conformation class 3 (Dataset-A) | | Descriptor: | CALCIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, Ryanodine receptor 1,Ryanodine receptor 1,RyR1, ... | | Authors: | Nayak, A.R, Samso, M. | | Deposit date: | 2021-12-31 | | Release date: | 2022-03-09 | | Last modified: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Ca 2+ -inactivation of the mammalian ryanodine receptor type 1 in a lipidic environment revealed by cryo-EM.
Elife, 11, 2022
|
|
7MBN
 
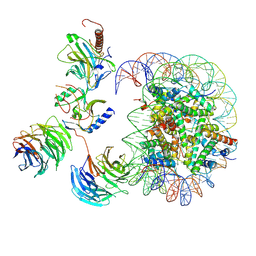 | | Cryo-EM structure of MLL1-NCP (H3K4M) complex, mode02 | | Descriptor: | DNA (146-MER), Histone H2A, Histone H2B 1.1, ... | | Authors: | Park, S.H, Ayoub, A, Lee, Y.T, Dou, Y, Cho, U. | | Deposit date: | 2021-04-01 | | Release date: | 2021-12-29 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY | | Cite: | Regulation of MLL1 Methyltransferase Activity in Two Distinct Nucleosome Binding Modes.
Biochemistry, 61, 2022
|
|
7MBM
 
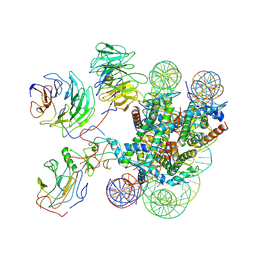 | | Cryo-EM structure of MLL1-NCP (H3K4M) complex, mode01 | | Descriptor: | DNA (145-MER), Histone H2A, Histone H2B 1.1, ... | | Authors: | Park, S.H, Ayoub, A, Lee, Y.T, Dou, Y, Cho, U. | | Deposit date: | 2021-04-01 | | Release date: | 2021-12-29 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY | | Cite: | Regulation of MLL1 Methyltransferase Activity in Two Distinct Nucleosome Binding Modes.
Biochemistry, 61, 2022
|
|
7M6A
 
 | | High resolution structure of the membrane embedded skeletal muscle ryanodine receptor | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CAFFEINE, CALCIUM ION, ... | | Authors: | Melville, Z, Kim, K, Clarke, O.B, Marks, A.R. | | Deposit date: | 2021-03-25 | | Release date: | 2021-09-08 | | Last modified: | 2022-01-19 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | High-resolution structure of the membrane-embedded skeletal muscle ryanodine receptor.
Structure, 30, 2022
|
|
