5E6U
 
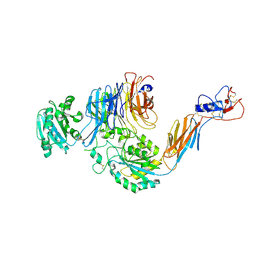 | | Structures of leukocyte integrin aLb2: The aI domain, the headpiece, and the pocket for the internal ligand | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Sen, M, Springer, T.A. | | Deposit date: | 2015-10-10 | | Release date: | 2016-04-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Leukocyte integrin alpha L beta 2 headpiece structures: The alpha I domain, the pocket for the internal ligand, and concerted movements of its loops.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5E6S
 
 | | Structures of leukocyte integrin aLB2: The aI domain, the headpiece, and the pocket for the internal ligand | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Integrin alpha-L, ... | | Authors: | Springer, T.A, Sen, M. | | Deposit date: | 2015-10-10 | | Release date: | 2016-03-02 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Leukocyte integrin alpha L beta 2 headpiece structures: The alpha I domain, the pocket for the internal ligand, and concerted movements of its loops.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5IDE
 
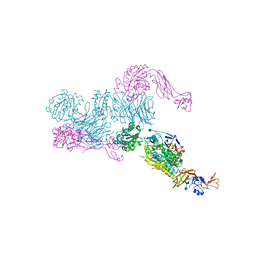
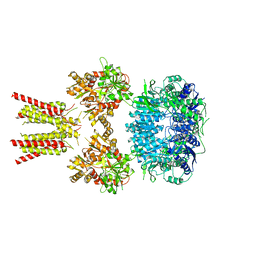 | | Cryo-EM structure of GluA2/3 AMPA receptor heterotetramer (model I) | | Descriptor: | Glutamate receptor 2, Glutamate receptor 3 | | Authors: | Herguedas, B, Garcia-Nafria, J, Fernandez-Leiro, R, Greger, I.H. | | Deposit date: | 2016-02-24 | | Release date: | 2016-03-16 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (8.25 Å) | | Cite: | Structure and organization of heteromeric AMPA-type glutamate receptors.
Science, 352, 2016
|
|
6T8Z
 
 | | NAD+-dependent fungal formate dehydrogenase from Chaetomium thermophilum: A ternary complex with the oxidised form of the cofactor NAD+ and the substrate formate both at a primary and secondary sites. | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, FORMIC ACID, ... | | Authors: | Isupov, M.N, Yelmazer, B, De Rose, S.A, Littlechild, J.A. | | Deposit date: | 2019-10-25 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Structural insights into the NAD + -dependent formate dehydrogenase mechanism revealed from the NADH complex and the formate NAD + ternary complex of the Chaetomium thermophilum enzyme.
J.Struct.Biol., 212, 2020
|
|
3PS9
 
 | | Crystal structure of MnmC from E. coli | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, S-ADENOSYLMETHIONINE, ... | | Authors: | Kim, J, Almo, S.C. | | Deposit date: | 2010-12-01 | | Release date: | 2010-12-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Structural basis for hypermodification of the wobble uridine in tRNA by bifunctional enzyme MnmC.
Bmc Struct.Biol., 13, 2013
|
|
1G3A
 
 | |
7Z1G
 
 | | Crystal structure of nonphosphorylated (Tyr216) GSK3b in complex with CX-4945 | | Descriptor: | 5-[(3-chlorophenyl)amino]benzo[c][2,6]naphthyridine-8-carboxylic acid, Glycogen synthase kinase-3 beta, IMIDAZOLE, ... | | Authors: | Grygier, P, Pustelny, K, Dubin, G, Czarna, A. | | Deposit date: | 2022-02-24 | | Release date: | 2023-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystal structure of nonphosphorylated (Tyr216) GSK3b kinase in complex with CX-4945
To Be Published
|
|
7Z1F
 
 | | Crystal structure of GSK3b in complex with CX-4945 | | Descriptor: | 5-[(3-chlorophenyl)amino]benzo[c][2,6]naphthyridine-8-carboxylic acid, Glycogen synthase kinase-3 beta, IMIDAZOLE, ... | | Authors: | Grygier, P, Pustelny, K, Dubin, G, Czarna, A. | | Deposit date: | 2022-02-24 | | Release date: | 2023-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of phosphorylated (Tyr216) GSK3b kinase in complex with CX-4945
To Be Published
|
|
7Z5N
 
 | | Crystal structure of DYRK1A in complex with CX-4945 | | Descriptor: | 1,2-ETHANEDIOL, 5-[(3-chlorophenyl)amino]benzo[c][2,6]naphthyridine-8-carboxylic acid, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Pustelny, K, Grygier, P, Golik, P, Dubin, G, Czarna, A. | | Deposit date: | 2022-03-09 | | Release date: | 2023-03-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Crystal structure DYRK1A in complex with CX-4945
To Be Published
|
|
1I00
 
 | | CRYSTAL STRUCTURE OF HUMAN THYMIDYLATE SYNTHASE, TERNARY COMPLEX WITH DUMP AND TOMUDEX | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, THYMIDYLATE SYNTHASE, TOMUDEX | | Authors: | Almog, R.A, Waddling, C.A, Maley, F, Maley, G.F, Van Roey, P. | | Deposit date: | 2001-01-27 | | Release date: | 2001-05-09 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a deletion mutant of human thymidylate synthase Delta (7-29) and its ternary complex with Tomudex and dUMP.
Protein Sci., 10, 2001
|
|
5AMK
 
 | | Cereblon isoform 4 from Magnetospirillum gryphiswaldense in multiple conformations, hexagonal crystal form | | Descriptor: | CEREBLON ISOFORM 4, DIMETHYL SULFOXIDE, S-Thalidomide, ... | | Authors: | Hartmann, M.D, Lupas, A.N, Hernandez Alvarez, B. | | Deposit date: | 2015-03-10 | | Release date: | 2015-07-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural Dynamics of the Cereblon Ligand Binding Domain.
Plos One, 10, 2015
|
|
5AMH
 
 | | Cereblon isoform 4 from Magnetospirillum gryphiswaldense in complex with Thalidomide, trigonal crystal form | | Descriptor: | CALCIUM ION, CEREBLON ISOFORM 4, CHLORIDE ION, ... | | Authors: | Hartmann, M.D, Lupas, A.N, Hernandez Alvarez, B. | | Deposit date: | 2015-03-10 | | Release date: | 2015-07-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural Dynamics of the Cereblon Ligand Binding Domain.
Plos One, 10, 2015
|
|
2J3J
 
 | | Crystal structure of Arabidopsis thaliana Double Bond Reductase (AT5G16970)-Ternary Complex I | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NADPH-dependent oxidoreductase 2-alkenal reductase | | Authors: | Youn, B, Kim, S.J, Moinuddin, S.G, Lee, C, Bedgar, D.L, Harper, A.R, Davin, L.B, Lewis, N.G, Kang, C. | | Deposit date: | 2006-08-21 | | Release date: | 2006-10-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Mechanistic and structural studies of apoform, binary, and ternary complexes of the Arabidopsis alkenal double bond reductase At5g16970.
J. Biol. Chem., 281, 2006
|
|
2OXC
 
 | | Human DEAD-box RNA helicase DDX20, DEAD domain in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Probable ATP-dependent RNA helicase DDX20 | | Authors: | Karlberg, T, Ogg, D, Arrowsmith, C.H, Berglund, H, Busam, R.D, Collins, R, Dahlgren, L.G, Edwards, A, Flodin, S, Flores, A, Graslund, S, Hallberg, B.M, Hammarstrom, M, Hogbom, M, Johansson, I, Kotenyova, T, Lehtio, L, Moche, M, Nordlund, P, Nyman, T, Persson, C, Sagemark, J, Stenmark, P, Sundstrom, M, Thorsell, A.G, Van Den Berg, S, Weigelt, J, Holmberg-Schiavone, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-02-20 | | Release date: | 2007-02-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Human DEAD-box RNA helicase DDX20
To be Published
|
|
2J3K
 
 | | Crystal structure of Arabidopsis thaliana Double Bond Reductase (AT5G16970)-Ternary Complex II | | Descriptor: | (2E,4R)-4-HYDROXYNON-2-ENAL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NADPH-dependent oxidoreductase 2-alkenal reductase | | Authors: | Youn, B, Kim, S.J, Moinuddin, S.G, Lee, C, Bedgar, D.L, Harper, A.R, Davin, L.B, Lewis, N.G, Kang, C. | | Deposit date: | 2006-08-22 | | Release date: | 2006-10-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Mechanistic and structural studies of apoform, binary, and ternary complexes of the Arabidopsis alkenal double bond reductase At5g16970.
J. Biol. Chem., 281, 2006
|
|
7UX4
 
 | |
5D6H
 
 | | Crystal structure of CsuC-CsuA/B chaperone-major subunit pre-assembly complex from Csu biofilm-mediating pili of Acinetobacter baumannii | | Descriptor: | CsuA/B, CsuC | | Authors: | Pakharukova, N.A, Tuitilla, M, Paavilainen, S, Zavialov, A. | | Deposit date: | 2015-08-12 | | Release date: | 2015-11-04 | | Last modified: | 2015-12-02 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Insight into Archaic and Alternative Chaperone-Usher Pathways Reveals a Novel Mechanism of Pilus Biogenesis.
Plos Pathog., 11, 2015
|
|
5DFK
 
 | |
5BU9
 
 | | Crystal structure of Beta-N-acetylhexosaminidase from Beutenbergia cavernae DSM 12333 | | Descriptor: | Beta-N-acetylhexosaminidase, GLYCEROL | | Authors: | Chang, C, Tan, K, Li, H, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-06-03 | | Release date: | 2015-06-17 | | Method: | X-RAY DIFFRACTION (2.255 Å) | | Cite: | Crystal structure of Beta-N-acetylhexosaminidase from Beutenbergia cavernae DSM 12333
To Be Published
|
|
5BKH
 
 | |
3MYW
 
 | | The Bowman-Birk type inhibitor from mung bean in ternary complex with porcine trypsin | | Descriptor: | Bowman-Birk type trypsin inhibitor, CALCIUM ION, Trypsin | | Authors: | Engh, R.A, Bode, W, Huber, R, Lin, G, Chi, C. | | Deposit date: | 2010-05-11 | | Release date: | 2010-12-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The 0.25-nm X-ray structure of the Bowman-Birk-type inhibitor from mung bean in ternary complex with porcine trypsin.
Eur.J.Biochem., 212, 1993
|
|
5AMJ
 
 | | Cereblon isoform 4 from Magnetospirillum gryphiswaldense in complex with Thalidomide, Wash II structure | | Descriptor: | CEREBLON ISOFORM 4, PHOSPHATE ION, S-Thalidomide, ... | | Authors: | Hartmann, M.D, Lupas, A.N, Hernandez Alvarez, B. | | Deposit date: | 2015-03-10 | | Release date: | 2015-07-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Dynamics of the Cereblon Ligand Binding Domain.
Plos One, 10, 2015
|
|
3PJP
 
 | | A Tandem SH2 Domain in Transcription Elongation Factor Spt6 Binds the Phosphorylated RNA Polymerase II C-terminal Repeat Domain(CTD) | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ACETATE ION, Transcription elongation factor SPT6 | | Authors: | Sun, M, Lariviere, L, Dengl, S, Mayer, A, Cramer, P. | | Deposit date: | 2010-11-10 | | Release date: | 2010-11-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A tandem SH2 domain in transcription elongation factor Spt6 binds the phosphorylated RNA polymerase II C-terminal repeat domain (CTD).
J.Biol.Chem., 285, 2010
|
|
5AMI
 
 | | Cereblon isoform 4 from Magnetospirillum gryphiswaldense in complex with Thalidomide, Wash I structure | | Descriptor: | CEREBLON ISOFORM 4, S-Thalidomide, ZINC ION | | Authors: | Hartmann, M.D, Lupas, A.N, Hernandez Alvarez, B. | | Deposit date: | 2015-03-10 | | Release date: | 2015-07-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Dynamics of the Cereblon Ligand Binding Domain.
Plos One, 10, 2015
|
|
3Q6F
 
 | | Crystal structure of Fab of human mAb 2909 specific for quaternary neutralizing epitope of HIV-1 gp120 | | Descriptor: | DI(HYDROXYETHYL)ETHER, Heavy chain of Fab of human mAb 2909, Light chain of Fab of human mAb 2909 | | Authors: | Spurrier, B, Sampson, J, Totrov, M, Li, H, O'Neal, T, William, C, Robinson, J, Gorny, M.K, Zolla-Pazner, S, Kong, X.P. | | Deposit date: | 2010-12-31 | | Release date: | 2011-05-25 | | Last modified: | 2011-10-12 | | Method: | X-RAY DIFFRACTION (3.192 Å) | | Cite: | Structural Analysis of Human and Macaque mAbs 2909 and 2.5B: Implications for the Configuration of the Quaternary Neutralizing Epitope of HIV-1 gp120.
Structure, 19, 2011
|
|
