8U1J
 
 | |
6S5F
 
 | | Structure of the human RAB39B in complex with GMPPNP | | Descriptor: | 1,2-ETHANEDIOL, GLYCINE, MAGNESIUM ION, ... | | Authors: | Diaz-Saez, L, Jung, S, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Huber, K, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-07-01 | | Release date: | 2020-07-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of the human RAB39B in complex with GMPPNP
To Be Published
|
|
6RWG
 
 | | Structure of HIV-1 CAcSP1NC mutant(W41A,M42A) interacting with maturation inhibitor EP39 | | Descriptor: | Gag polyprotein, ZINC ION | | Authors: | Chen, X, Coric, P, Larue, V, Nonin-Lecomte, S, Bouaziz, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-06-05 | | Release date: | 2020-05-20 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | The HIV-1 maturation inhibitor, EP39, interferes with the dynamic helix-coil equilibrium of the CA-SP1 junction of Gag.
Eur.J.Med.Chem., 204, 2020
|
|
5C40
 
 | | Crystal structure of human ribokinase in complex with AMPPCP in P21 spacegroup | | Descriptor: | PHOSPHATE ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, Ribokinase, ... | | Authors: | Park, J, Chakrabarti, J, Singh, B, Gupta, R.S, Junop, M.S. | | Deposit date: | 2015-06-17 | | Release date: | 2016-06-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of human ribokinase in complex with AMPPCP in P21 spacegroup
To Be Published
|
|
5CKS
 
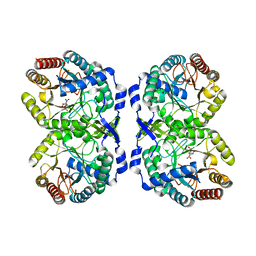 | | DAHP (3-deoxy-D-arabinoheptulosonate-7-phosphate) Synthase in complex with DAHP Oxime. | | Descriptor: | DAHP Oxime, Phospho-2-dehydro-3-deoxyheptonate aldolase, Phe-sensitive, ... | | Authors: | Berti, P, Junop, M, Balachandran, N. | | Deposit date: | 2015-07-15 | | Release date: | 2016-08-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1181 Å) | | Cite: | Potent Inhibition of 3-Deoxy-d-arabinoheptulosonate-7-phosphate (DAHP) Synthase by DAHP Oxime, a Phosphate Group Mimic.
Biochemistry, 55, 2016
|
|
8PY3
 
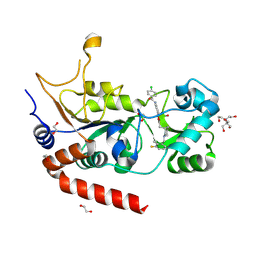 | | Crystal structure of human Sirt2 in complex with a 1,2,4-oxadiazole based inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-chloranyl-~{N}-[4-[5-[[(3~{S})-1-[(3-fluoranyl-2-methyl-phenyl)methyl]piperidin-3-yl]methyl]-1,2,4-oxadiazol-3-yl]phenyl]benzamide, ... | | Authors: | Friedrich, F, Colcerasa, A, Einsle, O, Jung, M. | | Deposit date: | 2023-07-24 | | Release date: | 2024-06-19 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure-Activity Studies of 1,2,4-Oxadiazoles for the Inhibition of the NAD + -Dependent Lysine Deacylase Sirtuin 2.
J.Med.Chem., 67, 2024
|
|
8G2Z
 
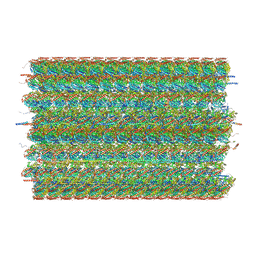 | | 48-nm doublet microtubule from Tetrahymena thermophila strain CU428 | | Descriptor: | B2B3_fMIP, B5B6_fMIP, CFAM166A, ... | | Authors: | Black, C.S, Kubo, S, Yang, S.K, Bui, K.H. | | Deposit date: | 2023-02-06 | | Release date: | 2023-06-14 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Native doublet microtubules from Tetrahymena thermophila reveal the importance of outer junction proteins.
Nat Commun, 14, 2023
|
|
8G3D
 
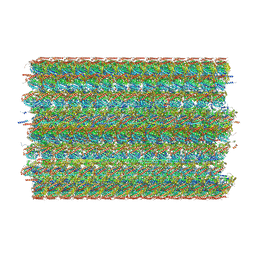 | | 48-nm doublet microtubule from Tetrahymena thermophila strain K40R | | Descriptor: | B2B3_fMIP, B5B6_fMIP, CFAM166A, ... | | Authors: | Black, C.S, Kubo, S, Yang, S.K, Bui, K.H. | | Deposit date: | 2023-02-07 | | Release date: | 2023-06-14 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Native doublet microtubules from Tetrahymena thermophila reveal the importance of outer junction proteins.
Nat Commun, 14, 2023
|
|
6TQD
 
 | | D00-D0 domain of Intimin | | Descriptor: | BROMIDE ION, CHLORIDE ION, Intimin, ... | | Authors: | Weikum, J, Leo, J.C, Morth, J.P. | | Deposit date: | 2019-12-16 | | Release date: | 2021-01-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | The extracellular juncture domains in the intimin passenger adopt a constitutively extended conformation inducing restraints to its sphere of action.
Sci Rep, 10, 2020
|
|
2WIW
 
 | | Crystal structures of Holliday junction resolvases from Archaeoglobus fulgidus bound to DNA substrate | | Descriptor: | 5'-D(*DC*DG*DG*DA*DT*DA*DT*DC*DC*DGP)-3', GLYCEROL, HEXANE-1,6-DIOL, ... | | Authors: | Carolis, C, Koehler, C, Sauter, C, Basquin, J, Suck, D, Toeroe, I. | | Deposit date: | 2009-05-18 | | Release date: | 2009-05-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structures of Holliday Junction Resolvases from Archaeoglobus Fulgidus Bound to DNA Substrate
To be Published
|
|
8OWZ
 
 | | Crystal structure of human Sirt2 in complex with a triazole-based SirReal | | Descriptor: | (R,R)-2,3-BUTANEDIOL, 1,2-ETHANEDIOL, 2-(4,6-dimethylpyrimidin-2-yl)sulfanyl-N-[5-[[3-[[1-(2-methoxyethyl)-1,2,3-triazol-4-yl]methoxy]phenyl]methyl]-1,3-thiazol-2-yl]ethanamide, ... | | Authors: | Friedrich, F, Zhang, L, Schiedel, M, Einsle, O, Jung, M. | | Deposit date: | 2023-04-28 | | Release date: | 2023-11-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Development of First-in-Class Dual Sirt2/HDAC6 Inhibitors as Molecular Tools for Dual Inhibition of Tubulin Deacetylation.
J.Med.Chem., 66, 2023
|
|
5LWQ
 
 | | CeuE (H227L variant) a periplasmic protein from Campylobacter jejuni | | Descriptor: | BROMIDE ION, Enterochelin uptake periplasmic binding protein, SODIUM ION | | Authors: | Wilde, E.J, Blagova, E, Hughes, A, Raines, D.J, Moroz, O.V, Turkenburg, J.P, Duhme-Klair, A.-K, Wilson, K.S. | | Deposit date: | 2016-09-19 | | Release date: | 2017-04-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Interactions of the periplasmic binding protein CeuE with Fe(III) n-LICAM(4-) siderophore analogues of varied linker length.
Sci Rep, 7, 2017
|
|
6GO3
 
 | | TdT chimera (Loop1 of pol mu) - apoenzyme | | Descriptor: | DNA nucleotidylexotransferase,DNA-directed DNA/RNA polymerase mu,DNA nucleotidylexotransferase | | Authors: | Loc'h, J, Gerodimos, C.A, Rosario, S, Lieber, M.R, Delarue, M. | | Deposit date: | 2018-06-01 | | Release date: | 2019-06-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural evidence for an intransbase selection mechanism involving Loop1 in polymerase mu at an NHEJ double-strand break junction.
J.Biol.Chem., 294, 2019
|
|
6GO4
 
 | | TdT chimera (Loop1 of pol mu) - binary complex with ddCTP | | Descriptor: | 2',3'-DIDEOXYCYTIDINE 5'-TRIPHOSPHATE, DNA nucleotidylexotransferase,DNA-directed DNA/RNA polymerase mu,DNA nucleotidylexotransferase, MAGNESIUM ION, ... | | Authors: | Loc'h, J, Gerodimos, C.A, Rosario, S, Lieber, M.R, Delarue, M. | | Deposit date: | 2018-06-01 | | Release date: | 2019-06-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural evidence for an intransbase selection mechanism involving Loop1 in polymerase mu at an NHEJ double-strand break junction.
J.Biol.Chem., 294, 2019
|
|
8WQ7
 
 | | Crystal structure of d(CGTATACG)2 with a four-carbon linker containing diacridine compound | | Descriptor: | DNA (5'-D(P*CP*GP*TP*AP*TP*AP*CP*G)-3'), MANGANESE (II) ION, N,N'-di(acridin-9-yl)butane-1,4-diamine | | Authors: | Huang, S.C, Satange, R.B, Hou, M.H. | | Deposit date: | 2023-10-11 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Targeting DNA junction sites by bis-intercalators induces topological changes with potent antitumor effects.
Nucleic Acids Res., 2024
|
|
8W7W
 
 | | Crystal structure of d(CGTATACG)2 with acridine complex | | Descriptor: | DNA (5'-D(P*CP*GP*TP*AP*TP*AP*CP*G)-3'), MAGNESIUM ION, ~{N},~{N}'-di(acridin-9-yl)pentane-1,5-diamine | | Authors: | Huang, S.C, Satange, R.B, Hou, M.H. | | Deposit date: | 2023-08-31 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Targeting DNA junction sites by bis-intercalators induces topological changes with potent antitumor effects.
Nucleic Acids Res., 2024
|
|
6GO7
 
 | | TdT chimera (Loop1 of pol mu) - full DNA synapsis complex | | Descriptor: | 2',3'-DIDEOXYCYTIDINE 5'-TRIPHOSPHATE, DNA (5'-D(*AP*AP*AP*AP*AP*C)-3'), DNA (5'-D(*TP*TP*TP*TP*TP*GP*C)-3'), ... | | Authors: | Loc'h, J, Gerodimos, C.A, Rosario, S, Lieber, M.R, Delarue, M. | | Deposit date: | 2018-06-01 | | Release date: | 2019-06-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural evidence for an intransbase selection mechanism involving Loop1 in polymerase mu at an NHEJ double-strand break junction.
J.Biol.Chem., 294, 2019
|
|
6TPL
 
 | | D0-D1 domain of Intimin | | Descriptor: | CHLORIDE ION, Intimin, MAGNESIUM ION | | Authors: | Weikum, J, Leo, J.C, Morth, J.P. | | Deposit date: | 2019-12-13 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The extracellular juncture domains in the intimin passenger adopt a constitutively extended conformation inducing restraints to its sphere of action.
Sci Rep, 10, 2020
|
|
3U4X
 
 | | Crystal structure of a lectin from Camptosema pedicellatum seeds in complex with 5-bromo-4-chloro-3-indolyl-alpha-D-mannose | | Descriptor: | 5-bromo-4-chloro-1H-indol-3-yl alpha-D-mannopyranoside, CALCIUM ION, Camptosema pedicellatum lectin (CPL), ... | | Authors: | Rocha, B.A.M, Teixeira, C.S, Moura, T.R, Silva, H.C, Pereira-Junior, F.N, Nagano, C.S, Delatorre, P, Cavada, B.S. | | Deposit date: | 2011-10-10 | | Release date: | 2012-09-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Crystal structure of the lectin of Camptosema pedicellatum: implications of a conservative substitution at the hydrophobic subsite.
J.Biochem., 152, 2012
|
|
6GO5
 
 | | TdT chimera (Loop1 of pol mu) - Ternary complex with 1-nt gapped DNA substrate | | Descriptor: | 2'-deoxy-5'-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]methyl}phosphoryl]cytidine, DNA (5'-D(*AP*CP*AP*GP*CP*G)-3'), DNA (5'-D(*CP*GP*CP*TP*GP*GP*CP*AP*AP*AP*CP*A)-3'), ... | | Authors: | Loc'h, J, Gerodimos, C.A, Rosario, S, Lieber, M.R, Delarue, M. | | Deposit date: | 2018-06-01 | | Release date: | 2019-06-05 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural evidence for an intransbase selection mechanism involving Loop1 in polymerase mu at an NHEJ double-strand break junction.
J.Biol.Chem., 294, 2019
|
|
1P27
 
 | | Crystal Structure of the Human Y14/Magoh complex | | Descriptor: | Mago nashi protein homolog, RNA-binding protein 8A | | Authors: | Lau, C.K, Diem, M.D, Dreyfuss, G, Van Duyne, G.D. | | Deposit date: | 2003-04-14 | | Release date: | 2003-08-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the y14-magoh core of the exon junction complex.
Curr.Biol., 13, 2003
|
|
7P1K
 
 | | Cryo EM structure of bison NHA2 in nano disc structure | | Descriptor: | CHOLESTEROL HEMISUCCINATE, Phosphatidylinositol, mitochondrial sodium/hydrogen exchanger 9B2 | | Authors: | Matsuoka, R, Fudim, R, Jung, S, Drew, D. | | Deposit date: | 2021-07-01 | | Release date: | 2022-01-26 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.64 Å) | | Cite: | Structure, mechanism and lipid-mediated remodeling of the mammalian Na + /H + exchanger NHA2.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7P1J
 
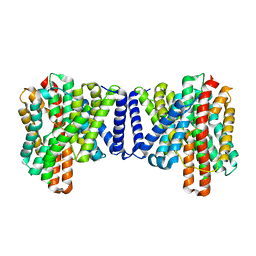 | | Cryo EM structure of bison NHA2 in detergent structure | | Descriptor: | mitochondrial sodium/hydrogen exchanger 9B2 | | Authors: | Matsuoka, R, Fudim, R, Jung, S, Drew, D. | | Deposit date: | 2021-07-01 | | Release date: | 2022-01-26 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Structure, mechanism and lipid-mediated remodeling of the mammalian Na + /H + exchanger NHA2.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7P1I
 
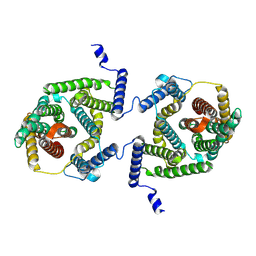 | | Cryo EM structure of bison NHA2 in detergent and N-terminal extension helix | | Descriptor: | mitochondrial sodium/hydrogen exchanger 9B2 | | Authors: | Matsuoka, R, Fudim, R, Jung, S, Drew, D. | | Deposit date: | 2021-07-01 | | Release date: | 2022-01-26 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Structure, mechanism and lipid-mediated remodeling of the mammalian Na + /H + exchanger NHA2.
Nat.Struct.Mol.Biol., 29, 2022
|
|
6PPW
 
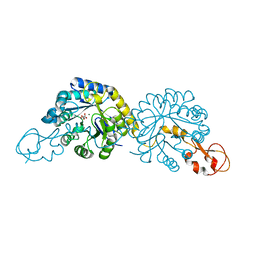 | | Crystal structure of NeuB, an N-acetylneuraminate synthase from Neisseria meningitidis, in complex with magnesium and malate | | Descriptor: | D-MALATE, MAGNESIUM ION, N-acetylneuraminate synthase | | Authors: | Rosanally, A.Z, Junop, M.S, Berti, P.J. | | Deposit date: | 2019-07-08 | | Release date: | 2019-10-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | NeuNAc Oxime: A Slow-Binding and Effectively Irreversible Inhibitor of the Sialic Acid Synthase NeuB.
Biochemistry, 58, 2019
|
|
