8F2S
 
 | | Cryo-EM structure of Torpedo nicotinic acetylcholine receptor in complex with rocuronium, pore-blocked state | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Goswami, U, Rahman, M.M, Teng, J, Hibbs, R.E. | | Deposit date: | 2022-11-08 | | Release date: | 2023-06-07 | | Last modified: | 2023-06-14 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural interplay of anesthetics and paralytics on muscle nicotinic receptors.
Nat Commun, 14, 2023
|
|
6RZ8
 
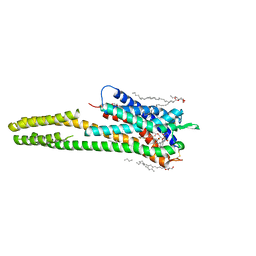 | | Crystal structure of the human cysteinyl leukotriene receptor 2 in complex with ONO-2080365 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2~{S})-8-[[4-[4-[2,3-bis(fluoranyl)phenoxy]butoxy]-2-fluoranyl-phenyl]carbonylamino]-4-(4-oxidanyl-4-oxidanylidene-but yl)-2,3-dihydro-1,4-benzoxazine-2-carboxylic acid, Cysteinyl leukotriene receptor 2,Soluble cytochrome b562,Cysteinyl leukotriene receptor 2, ... | | Authors: | Gusach, A, Luginina, A, Marin, E, Brouillette, R.L, Besserer-Offroy, E, Longpre, J.M, Ishchenko, A, Popov, P, Fujimoto, T, Maruyama, T, Stauch, B, Ergasheva, M, Romanovskaya, D, Stepko, A, Kovalev, K, Shevtsov, M, Gordeliy, V, Han, G.W, Sarret, P, Katritch, V, Borshchevskiy, V, Mishin, A, Cherezov, V. | | Deposit date: | 2019-06-12 | | Release date: | 2019-12-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of ligand selectivity and disease mutations in cysteinyl leukotriene receptors.
Nat Commun, 10, 2019
|
|
6TW0
 
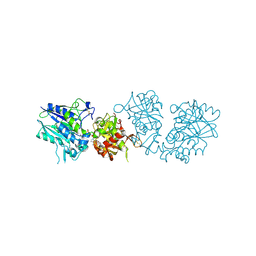 | | Human CD73 (ecto 5'-nucleotidase) in complex with PSB12690 (an AOPCP derivative, compound 10 in publication) in the closed state | | Descriptor: | 5'-nucleotidase, ZINC ION, [[(2~{R},3~{S},4~{R},5~{R})-5-(6-azanyl-2-oxidanylidene-3~{H}-purin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl]methylphosphonic acid | | Authors: | Pippel, J, Strater, N. | | Deposit date: | 2020-01-10 | | Release date: | 2020-02-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | 2-Substituted alpha , beta-Methylene-ADP Derivatives: Potent Competitive Ecto-5'-nucleotidase (CD73) Inhibitors with Variable Binding Modes.
J.Med.Chem., 63, 2020
|
|
1AZB
 
 | |
3NM1
 
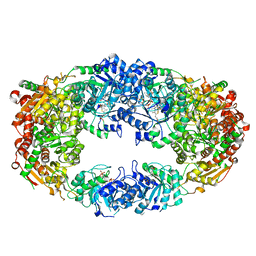 | | The Crystal Structure of Candida glabrata THI6, a Bifunctional Enzyme involved in Thiamin Biosyhthesis of Eukaryotes | | Descriptor: | 2-TRIFLUOROMETHYL-5-METHYLENE-5H-PYRIMIDIN-4-YLIDENEAMINE, 4-methyl-5-[2-(phosphonooxy)ethyl]-1,3-thiazole-2-carboxylic acid, MAGNESIUM ION, ... | | Authors: | Paul, D, Chatterjee, A, Begley, T.P, Ealick, S.E. | | Deposit date: | 2010-06-21 | | Release date: | 2010-11-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.211 Å) | | Cite: | Domain Organization in Candida glabrata THI6, a Bifunctional Enzyme Required for Thiamin Biosynthesis in Eukaryotes .
Biochemistry, 49, 2010
|
|
6TWF
 
 | | Human CD73 (ecto 5'-nucleotidase) in complex with PSB12604 (an AOPCP derivative, compound 21 in publication) in the closed state | | Descriptor: | 5'-nucleotidase, CALCIUM ION, ZINC ION, ... | | Authors: | Pippel, J, Strater, N. | | Deposit date: | 2020-01-13 | | Release date: | 2020-02-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | 2-Substituted alpha , beta-Methylene-ADP Derivatives: Potent Competitive Ecto-5'-nucleotidase (CD73) Inhibitors with Variable Binding Modes.
J.Med.Chem., 63, 2020
|
|
8F5F
 
 | | human branched chain ketoacid dehydrogenase kinase in complex with inhibitors | | Descriptor: | (2P)-2-[(4P)-4-{6-[(1-ethylcyclopropyl)methoxy]pyridin-3-yl}-1,3-thiazol-2-yl]benzoic acid, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Liu, S, Roth Flach, R, Bollinger, E, Filipski, K. | | Deposit date: | 2022-11-14 | | Release date: | 2023-07-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.149 Å) | | Cite: | Small molecule branched-chain ketoacid dehydrogenase kinase (BDK) inhibitors with opposing effects on BDK protein levels.
Nat Commun, 14, 2023
|
|
5LM0
 
 | | NMR spatial structure of Tk-hefu peptide | | Descriptor: | L-2 | | Authors: | Mineev, K.S, Berkut, A.A, Novikova, E.V, Oparin, P.B, Grishin, E.V, Arseniev, A.S, Vassilevski, A.A. | | Deposit date: | 2016-07-28 | | Release date: | 2017-08-16 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Protein Surface Topography as a tool to enhance the selective activity of a potassium channel blocker.
J.Biol.Chem., 2019
|
|
6ZOI
 
 | | A lid blocking mechanism of a cone snail toxin revealed at the atomic level | | Descriptor: | Conknunitzin-C3 mutante | | Authors: | Saikia, C, Altman-Gueta, H, Dym, O, Frolow, F, Gurevitz, M, Gordon, D, Reuveny, E, Karbat, I. | | Deposit date: | 2020-07-07 | | Release date: | 2021-07-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.798 Å) | | Cite: | A lid blocking mechanism of a cone snail toxin revealed at the atomic level
To Be Published
|
|
8TNR
 
 | | Cryo-EM structure of DDB1dB:CRBN:PT-179:SD40, conformation 2 | | Descriptor: | 2-[(3S)-2,6-dioxopiperidin-3-yl]-5-(morpholin-4-yl)-1H-isoindole-1,3(2H)-dione, DNA damage-binding protein 1, Maltose/maltodextrin-binding periplasmic protein,SD40, ... | | Authors: | Roy Burman, S.S, Hunkeler, M, Fischer, E.S. | | Deposit date: | 2023-08-02 | | Release date: | 2024-03-13 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Continuous evolution of compact protein degradation tags regulated by selective molecular glues.
Science, 383, 2024
|
|
5LME
 
 | | Specific-DNA binding activity of the cross-brace zinc finger motif of the piggyBac transposase | | Descriptor: | ZINC ION, piggyBac transposase | | Authors: | Morellet, N, Taylor, J.A, Wieninger, S, Moriau, S, Li, X, Lescop, E, Mathy, N, Bischerour, J, Betermier, M, Bardiaux, B, Nilges, M, Craig, N.L, Hickman, A.B, Dyda, F, Guittet, E. | | Deposit date: | 2016-07-30 | | Release date: | 2017-12-20 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Sequence-specific DNA binding activity of the cross-brace zinc finger motif of the piggyBac transposase.
Nucleic Acids Res., 46, 2018
|
|
2IH4
 
 | |
6TZI
 
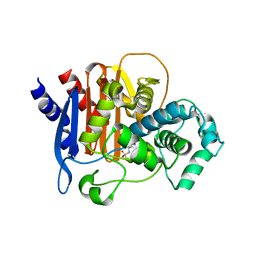 | | ADC-7 in complex with boronic acid transition state inhibitor PFC_001 | | Descriptor: | Beta-lactamase, phosphonooxy-[[4-[[2,2,2-tris(fluoranyl)ethylsulfonylamino]methyl]-1,2,3-triazol-1-yl]methyl]borinic acid | | Authors: | Fish, E.R, Powers, R.A, Wallar, B.J. | | Deposit date: | 2019-08-12 | | Release date: | 2020-06-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.744 Å) | | Cite: | 1,2,3-Triazolylmethaneboronate: A Structure Activity Relationship Study of a Class of beta-Lactamase Inhibitors againstAcinetobacter baumanniiCephalosporinase.
Acs Infect Dis., 6, 2020
|
|
3N7U
 
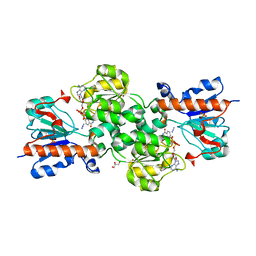 | | NAD-dependent formate dehydrogenase from higher-plant Arabidopsis thaliana in complex with NAD and azide | | Descriptor: | AZIDE ION, Formate dehydrogenase, GLYCEROL, ... | | Authors: | Shabalin, I.G, Polyakov, K.M, Serov, A.E, Skirgello, O.E, Sadykhov, E.G, Dorovatovskiy, P.V, Tishkov, V.I, Popov, V.O. | | Deposit date: | 2010-05-27 | | Release date: | 2010-06-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of the apo and holo forms of NAD-dependent formate dehydrogenase from the higher-plant Arabidopsis Thaliana
To be Published
|
|
1F4B
 
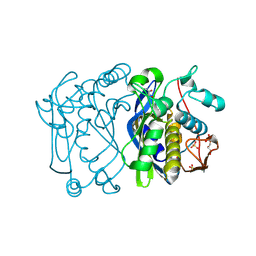 | | CRYSTAL STRUCTURE OF ESCHERICHIA COLI THYMIDYLATE SYNTHASE | | Descriptor: | GLYCEROL, SULFATE ION, THYMIDYLATE SYNTHASE | | Authors: | Erlanson, D.A, Braisted, A.C, Raphael, D.R, Randal, M, Stroud, R.M, Gordon, E, Wells, J.A. | | Deposit date: | 2000-06-07 | | Release date: | 2000-06-22 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Site-directed ligand discovery.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
8F5J
 
 | | human branched chain ketoacid dehydrogenase kinase in complex with inhibitors | | Descriptor: | 3-chloro-5-fluorothieno[3,2-b]thiophene-2-carboxylic acid, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Liu, S, Roth Flach, R, Bollinger, E, Filipski, K. | | Deposit date: | 2022-11-14 | | Release date: | 2023-07-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.545 Å) | | Cite: | Small molecule branched-chain ketoacid dehydrogenase kinase (BDK) inhibitors with opposing effects on BDK protein levels.
Nat Commun, 14, 2023
|
|
6TZF
 
 | | ADC-7 in complex with boronic acid transition state inhibitor S17079 | | Descriptor: | 3-(1-{[hydroxy(phosphonooxy)boranyl]methyl}-1H-1,2,3-triazol-4-yl)benzoic acid, Beta-lactamase | | Authors: | Fish, E.R, Powers, R.A, Wallar, B.J. | | Deposit date: | 2019-08-12 | | Release date: | 2020-06-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | 1,2,3-Triazolylmethaneboronate: A Structure Activity Relationship Study of a Class of beta-Lactamase Inhibitors againstAcinetobacter baumanniiCephalosporinase.
Acs Infect Dis., 6, 2020
|
|
7JXU
 
 | | Structure of monobody 32 human MLKL pseudokinase domain complex | | Descriptor: | 1,2-ETHANEDIOL, Mixed lineage kinase domain-like protein, Monobody 32 | | Authors: | Meng, Y, Garnish, S.E, Koide, A, Koide, S, Czabotar, P.E, Murphy, J.M. | | Deposit date: | 2020-08-28 | | Release date: | 2021-03-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Conformational interconversion of MLKL and disengagement from RIPK3 precede cell death by necroptosis.
Nat Commun, 12, 2021
|
|
1IM3
 
 | | Crystal Structure of the human cytomegalovirus protein US2 bound to the MHC class I molecule HLA-A2/tax | | Descriptor: | HLA CLASS I HISTOCOMPATIBILITY ANTIGEN, A-2 ALPHA CHAIN, Human T-cell lymphotropic virus type 1 Tax peptide, ... | | Authors: | Gewurz, B.E, Gaudet, R, Tortorella, D, Wang, E.W, Ploegh, H.L, Wiley, D.C. | | Deposit date: | 2001-05-09 | | Release date: | 2001-06-06 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Antigen presentation subverted: Structure of the human cytomegalovirus protein US2 bound to the class I molecule HLA-A2.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
5EUQ
 
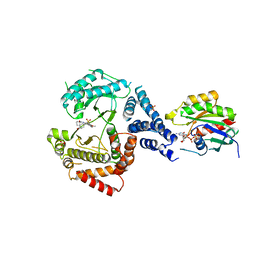 | |
8F5S
 
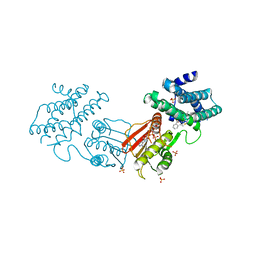 | | human branched chain ketoacid dehydrogenase kinase in complex with inhibitors | | Descriptor: | (2M)-2-[2-(4-methylphenyl)-1,3-thiazol-4-yl]benzoic acid, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Liu, S, Roth Flach, R, Bollinger, E, Filipski, K. | | Deposit date: | 2022-11-15 | | Release date: | 2023-07-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.793 Å) | | Cite: | Small molecule branched-chain ketoacid dehydrogenase kinase (BDK) inhibitors with opposing effects on BDK protein levels.
Nat Commun, 14, 2023
|
|
5MCF
 
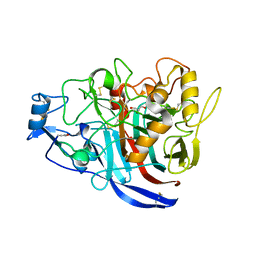 | | Radiation damage to GH7 Family Cellobiohydrolase from Daphnia pulex: Dose (DWD) 7.59 MGy | | Descriptor: | Cellobiohydrolase CHBI, GLYCEROL, SULFATE ION | | Authors: | Bury, C.S, McGeehan, J.E, Ebrahim, A, Garman, E.F. | | Deposit date: | 2016-11-09 | | Release date: | 2017-01-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | OH cleavage from tyrosine: debunking a myth.
J Synchrotron Radiat, 24, 2017
|
|
8CBR
 
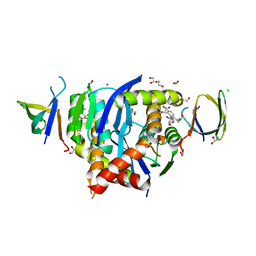 | | HIV-1 Integrase Catalytic Core Domain and C-Terminal Domain in Complex with Allosteric Integrase Inhibitor BDM-2 | | Descriptor: | (2S)-2-[3-cyclopropyl-2-(3,4-dihydro-2H-chromen-6-yl)-6-methyl-phenyl]-2-[(2-methylpropan-2-yl)oxy]ethanoic acid, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Singer, M.R, Pye, V.E, Yu, Z, Cherepanov, P. | | Deposit date: | 2023-01-25 | | Release date: | 2023-06-07 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Biological and Structural Analyses of New Potent Allosteric Inhibitors of HIV-1 Integrase.
Antimicrob.Agents Chemother., 67, 2023
|
|
8CBU
 
 | | HIV-1 Integrase Catalytic Core Domain and C-Terminal Domain in Complex with Allosteric Integrase Inhibitor MUT884 | | Descriptor: | (2S)-2-[3-cyclopropyl-6-methyl-2-(5-methyl-3,4-dihydro-2H-chromen-6-yl)phenyl]-2-cyclopropyloxy-ethanoic acid, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Singer, M.R, Pye, V.E, Yu, Z, Cherepanov, P. | | Deposit date: | 2023-01-25 | | Release date: | 2023-06-07 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Biological and Structural Analyses of New Potent Allosteric Inhibitors of HIV-1 Integrase.
Antimicrob.Agents Chemother., 67, 2023
|
|
8CBV
 
 | | HIV-1 Integrase Catalytic Core Domain and C-Terminal Domain in Complex with Allosteric Integrase Inhibitor MUT916 | | Descriptor: | (2~{S})-2-[3-cyclopropyl-2-(8-fluoranyl-5-methyl-3,4-dihydro-2~{H}-chromen-6-yl)-6-methyl-phenyl]-2-cyclopropyloxy-ethanoic acid, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Singer, M.R, Pye, V.E, Yu, Z, Cherepanov, P. | | Deposit date: | 2023-01-25 | | Release date: | 2023-06-07 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Biological and Structural Analyses of New Potent Allosteric Inhibitors of HIV-1 Integrase.
Antimicrob.Agents Chemother., 67, 2023
|
|
