4XE9
 
 | |
8YE6
 
 | |
8YE9
 
 | |
7UBA
 
 | | Structure of fungal Hop1 CBR domain | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, HORMA domain-containing protein, PENTAETHYLENE GLYCOL, ... | | Authors: | Ur, S.N, Corbett, K.D. | | Deposit date: | 2022-03-14 | | Release date: | 2023-03-29 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.599 Å) | | Cite: | Chromatin binding by HORMAD proteins regulates meiotic recombination initiation.
Embo J., 43, 2024
|
|
4Q2W
 
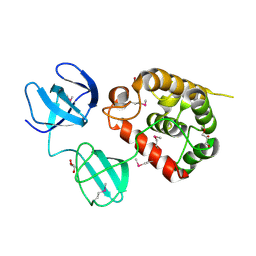 | | Crystal Structure of pneumococcal peptidoglycan hydrolase LytB | | Descriptor: | GLYCEROL, Putative endo-beta-N-acetylglucosaminidase | | Authors: | Bai, X.H, Chen, H.J, Jiang, Y.L, Wen, Z, Cheng, W, Li, Q, Zhang, J.R, Chen, Y, Zhou, C.Z. | | Deposit date: | 2014-04-10 | | Release date: | 2014-07-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure of pneumococcal peptidoglycan hydrolase LytB reveals insights into the bacterial cell wall remodeling and pathogenesis.
J.Biol.Chem., 289, 2014
|
|
8PM4
 
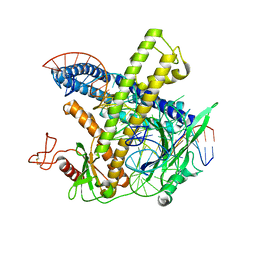 | | Cryo-EM structure of the Cas12m-crRNA-target DNA complex | | Descriptor: | DNA oligoduplex, non-target strand, chain D, ... | | Authors: | Sasnauskas, G, Tamulaitiene, G, Karvelis, T, Bigelyte, G, Siksnys, V. | | Deposit date: | 2023-06-28 | | Release date: | 2024-02-07 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Innate programmable DNA binding by CRISPR-Cas12m effectors enable efficient base editing.
Nucleic Acids Res., 52, 2024
|
|
4UN3
 
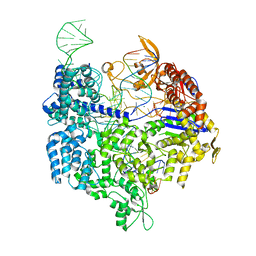 | | Crystal structure of Cas9 bound to PAM-containing DNA target | | Descriptor: | CRISPR-ASSOCIATED ENDONUCLEASE CAS9/CSN1, MAGNESIUM ION, NON-TARGET DNA STRAND, ... | | Authors: | Anders, C, Niewoehner, O, Duerst, A, Jinek, M. | | Deposit date: | 2014-05-25 | | Release date: | 2014-07-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.593 Å) | | Cite: | Structural Basis of Pam-Dependent Target DNA Recognition by the Cas9 Endonuclease
Nature, 513, 2014
|
|
4UN5
 
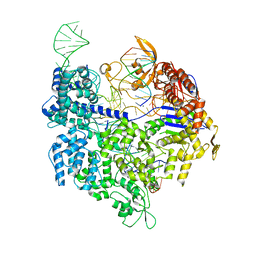 | | Crystal structure of Cas9 bound to PAM-containing DNA target containing mismatches at positions 1-3 | | Descriptor: | CRISPR-ASSOCIATED ENDONUCLEASE CAS9/CSN1, MAGNESIUM ION, NON-TARGET DNA STRAND, ... | | Authors: | Anders, C, Niewoehner, O, Duerst, A, Jinek, M. | | Deposit date: | 2014-05-25 | | Release date: | 2014-07-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis of Pam-Dependent Target DNA Recognition by the Cas9 Endonuclease
Nature, 513, 2014
|
|
4UN4
 
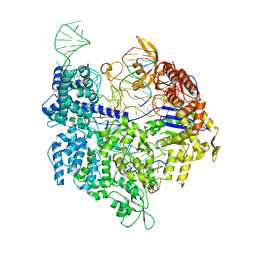 | | Crystal structure of Cas9 bound to PAM-containing DNA target with mismatches at positions 1-2 | | Descriptor: | CRISPR-ASSOCIATED ENDONUCLEASE CAS9/CSN1, MAGNESIUM ION, NON-TARGET DNA STRAND, ... | | Authors: | Anders, C, Niewoehner, O, Duerst, A, Jinek, M. | | Deposit date: | 2014-05-25 | | Release date: | 2014-07-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.371 Å) | | Cite: | Structural Basis of Pam-Dependent Target DNA Recognition by the Cas9 Endonuclease
Nature, 513, 2014
|
|
3L8C
 
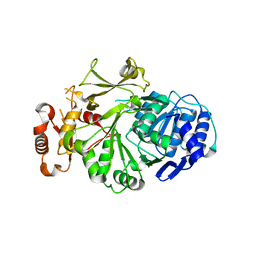 | |
3LGX
 
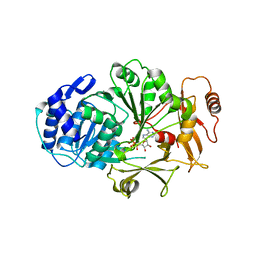 | |
8DG4
 
 | | Group A streptococcus Enolase K252A, K255A, K434A, K435A mutant | | Descriptor: | Enolase | | Authors: | Tjia-Fleck, S.C, Readnour, B.M, Castellino, F.J. | | Deposit date: | 2022-06-23 | | Release date: | 2022-12-14 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | High-Resolution Single-Particle Cryo-EM Hydrated Structure of Streptococcus pyogenes Enolase Offers Insights into Its Function as a Plasminogen Receptor.
Biochemistry, 62, 2023
|
|
6O0X
 
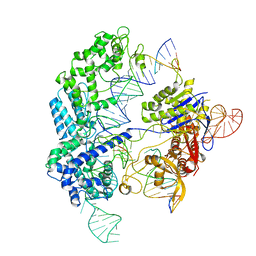 | | Conformational states of Cas9-sgRNA-DNA ternary complex in the presence of magnesium | | Descriptor: | 3' product of target strand DNA, 5' product of target strand DNA, CRISPR-associated endonuclease Cas9/Csn1, ... | | Authors: | Zhu, X, Clarke, R, Puppala, A.K, Chittori, S, Merk, A, Merrill, B.J, Simonovic, M, Subramaniam, S. | | Deposit date: | 2019-02-17 | | Release date: | 2019-07-10 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Cryo-EM structures reveal coordinated domain motions that govern DNA cleavage by Cas9.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6NKJ
 
 | | 1.3 Angstrom Resolution Crystal Structure of UDP-N-acetylglucosamine 1-carboxyvinyltransferase from Streptococcus pneumoniae in Complex with (2R)-2-(phosphonooxy)propanoic acid. | | Descriptor: | (2R)-2-(phosphonooxy)propanoic acid, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Kiryukhina, O, Grimshaw, S, Kwon, K, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-01-07 | | Release date: | 2019-01-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | 1.3 Angstrom Resolution Crystal Structure of UDP-N-acetylglucosamine 1-carboxyvinyltransferase from Streptococcus pneumoniae in Complex with (2R)-2-(phosphonooxy)propanoic acid.
To Be Published
|
|
6MK7
 
 | | Solution structure of the large extracellular loop of FtsX in Streptococcus pneumoniae | | Descriptor: | Cell division protein FtsX | | Authors: | Edmonds, K.A, Fu, Y, Wu, H, Rued, B.E, Bruce, K.E, Winkler, M.E, Giedroc, D.P. | | Deposit date: | 2018-09-25 | | Release date: | 2019-02-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of the Large Extracellular Loop of FtsX and Its Interaction with the Essential Peptidoglycan Hydrolase PcsB in Streptococcus pneumoniae.
MBio, 10, 2019
|
|
5EEH
 
 | | Crystal structure of carminomycin-4-O-methyltransferase DnrK in complex with SAH and 2-chloro-4-nitrophenol | | Descriptor: | 2-chloranyl-4-nitro-phenol, Carminomycin 4-O-methyltransferase DnrK, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Wang, F, Singh, S, Thorson, J.S, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2015-10-22 | | Release date: | 2015-12-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Functional AdoMet Isosteres Resistant to Classical AdoMet Degradation Pathways.
Acs Chem.Biol., 11, 2016
|
|
5EEG
 
 | | Crystal structure of carminomycin-4-O-methyltransferase DnrK in complex with tetrazole-SAH | | Descriptor: | (2~{R},3~{R},4~{S},5~{S})-2-(6-aminopurin-9-yl)-5-[[(3~{S})-3-azanyl-3-(1~{H}-1,2,3,4-tetrazol-5-yl)propyl]sulfanylmethyl]oxolane-3,4-diol, Carminomycin 4-O-methyltransferase DnrK | | Authors: | Wang, F, Singh, S, Thorson, J.S, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2015-10-22 | | Release date: | 2015-12-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.255 Å) | | Cite: | Functional AdoMet Isosteres Resistant to Classical AdoMet Degradation Pathways.
Acs Chem.Biol., 11, 2016
|
|
3MHF
 
 | | Tagatose-1,6-bisphosphate aldolase from Streptococcus pyogenes | | Descriptor: | CALCIUM ION, Tagatose 1,6-diphosphate aldolase 2 | | Authors: | LowKam, C. | | Deposit date: | 2010-04-07 | | Release date: | 2010-04-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structure of a class I tagatose-1,6-bisphosphate aldolase: investigation into an apparent loss of stereospecificity.
J.Biol.Chem., 285, 2010
|
|
3E0M
 
 | | Crystal structure of fusion protein of MsrA and MsrB | | Descriptor: | Peptide methionine sulfoxide reductase msrA/msrB 1, Short peptide SHMAEI | | Authors: | Kim, Y.K, Hwang, K.Y. | | Deposit date: | 2008-07-31 | | Release date: | 2009-06-16 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and Kinetic Analysis of an MsrA-MsrB Fusion Protein from Streptococcus pneumoniae
Mol.Microbiol., 72, 2009
|
|
3JRK
 
 | | A putative tagatose 1,6-diphosphate aldolase from Streptococcus pyogenes | | Descriptor: | GLYCEROL, Tagatose 1,6-diphosphate aldolase 2 | | Authors: | Fan, Y, Volkart, L, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-09-08 | | Release date: | 2009-09-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | The crystal structure of a putative tagatose 1,6-diphosphate aldolase from Streptococcus pyogenes
To be Published
|
|
6O0Z
 
 | | Conformational states of Cas9-sgRNA-DNA ternary complex in the presence of magnesium | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, non-target strand DNA, single guide RNA, ... | | Authors: | Zhu, X, Clarke, R, Puppala, A.K, Chittori, S, Merk, A, Merrill, B.J, Simonovic, M, Subramaniam, S. | | Deposit date: | 2019-02-17 | | Release date: | 2019-07-10 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures reveal coordinated domain motions that govern DNA cleavage by Cas9.
Nat.Struct.Mol.Biol., 26, 2019
|
|
5CTV
 
 | | Catalytic domain of LytA, the major autolysin of Streptococcus pneumoniae, (C60A, H133A, C136A mutant) complexed with peptidoglycan fragment | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-methyl 2-acetamido-3-O-[(1R)-1-carboxyethyl]-2-deoxy-beta-D-glucopyranoside-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-methyl 2-acetamido-3-O-[(1R)-1-carboxyethyl]-2-deoxy-beta-D-glucopyranoside, Autolysin, fragment of peptidoglycan | | Authors: | Achour, A, Sandalova, T, Mellroth, P. | | Deposit date: | 2015-07-24 | | Release date: | 2016-06-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | The crystal structure of the major pneumococcal autolysin LytA in complex with a large peptidoglycan fragment reveals the pivotal role of glycans for lytic activity.
Mol.Microbiol., 101, 2016
|
|
4IVV
 
 | |
5F2M
 
 | |
5F2I
 
 | |
