5Y2G
 
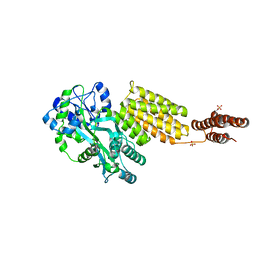 | | Structure of MBP tagged GBS CAMP | | Descriptor: | Maltose-binding periplasmic protein,Protein B, SULFATE ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Jin, T, Li, Y. | | Deposit date: | 2017-07-25 | | Release date: | 2019-02-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure determination of the CAMP factor of Streptococcus agalactiae with the aid of an MBP tag and insights into membrane-surface attachment.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
1WE0
 
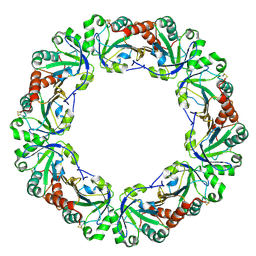 | | Crystal structure of peroxiredoxin (AhpC) from Amphibacillus xylanus | | Descriptor: | AMMONIUM ION, alkyl hydroperoxide reductase C | | Authors: | Kitano, K, Kita, A, Hakoshima, T, Niimura, Y, Miki, K. | | Deposit date: | 2004-05-21 | | Release date: | 2005-03-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of decameric peroxiredoxin (AhpC) from Amphibacillus xylanus
Proteins, 59, 2005
|
|
5WK7
 
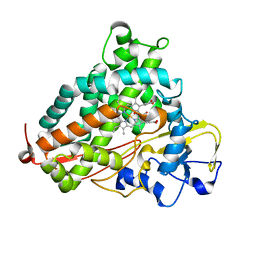 | | P450cam mutant R186A | | Descriptor: | 5-EXO-HYDROXYCAMPHOR, Camphor 5-monooxygenase, POTASSIUM ION, ... | | Authors: | Batabyal, D, Poulos, T.L. | | Deposit date: | 2017-07-24 | | Release date: | 2017-09-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.983 Å) | | Cite: | Effect of Redox Partner Binding on Cytochrome P450 Conformational Dynamics.
J. Am. Chem. Soc., 139, 2017
|
|
1FCO
 
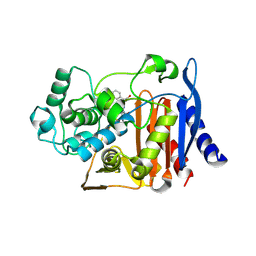 | | CRYSTAL STRUCTURE OF THE E. COLI AMPC BETA-LACTAMASE COVALENTLY ACYLATED WITH THE INHIBITORY BETA-LACTAM, MOXALACTAM | | Descriptor: | (2R)-2-[(1R)-1-{[(2S)-2-carboxy-2-(4-hydroxyphenyl)acetyl]amino}-1-methoxy-2-oxoethyl]-5-methylidene-5,6-dihydro-2H-1,3-oxazine-4-carboxylic acid, BETA-LACTAMASE | | Authors: | Patera, A, Blaszczak, L.C, Shoichet, B.K. | | Deposit date: | 2000-07-19 | | Release date: | 2000-12-04 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structures of Substrate and Inhibitor Complexes with AmpC -Lactamase: Possible Implications for Substrate-Assisted Catalysis
J.Am.Chem.Soc., 122, 2000
|
|
1FCN
 
 | |
6B76
 
 | | Crystal Structure of human NAMPT in complex with NVP-LVR596 | | Descriptor: | (1S,2S)-N-{4-[(1S)-1-(propanoylamino)ethyl]phenyl}-2-(pyridin-3-yl)cyclopropane-1-carboxamide, Nicotinamide phosphoribosyltransferase, PHOSPHATE ION | | Authors: | Weihofen, W.A, Thigale, S. | | Deposit date: | 2017-10-03 | | Release date: | 2018-10-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Identification and structure based design of cellularly active cyclo-propyl carboxamide Nicotinamide phosphoribosyltransferase (NAMPT) inhibitors
To Be Published
|
|
1FCM
 
 | |
8S62
 
 | |
3TSN
 
 | | 4-hydroxythreonine-4-phosphate dehydrogenase from Campylobacter jejuni | | Descriptor: | 4-hydroxythreonine-4-phosphate dehydrogenase, NICKEL (II) ION, UNKNOWN LIGAND | | Authors: | Osipiuk, J, Gu, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-09-13 | | Release date: | 2011-10-12 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | 4-hydroxythreonine-4-phosphate dehydrogenase from Campylobacter jejuni.
To be Published
|
|
3LXI
 
 | | Crystal Structure of Camphor-Bound CYP101D1 | | Descriptor: | CAMPHOR, Cytochrome P450, PHOSPHATE ION, ... | | Authors: | Yang, W, Bell, S.G, Wang, H, Bartlam, M, Wong, L.L, Rao, Z. | | Deposit date: | 2010-02-25 | | Release date: | 2010-06-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular characterization of a class I P450 electron transfer system from Novosphingobium aromaticivorans DSM12444
J.Biol.Chem., 285, 2010
|
|
5F1U
 
 | | biomimetic design results in a potent allosteric inhibitor of dihydrodipicolinate synthase from Campylobacter jejuni | | Descriptor: | (2R,5R)-2,5-diamino-2,5-bis(4-aminobutyl)hexanedioic acid, 1,2-ETHANEDIOL, 4-hydroxy-tetrahydrodipicolinate synthase, ... | | Authors: | Conly, C.J.T, Palmer, D.R.J, Sanders, D.A.R. | | Deposit date: | 2015-11-30 | | Release date: | 2016-02-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Biomimetic Design Results in a Potent Allosteric Inhibitor of Dihydrodipicolinate Synthase from Campylobacter jejuni.
J.Am.Chem.Soc., 138, 2016
|
|
3N0L
 
 | | Crystal structure of serine hydroxymethyltransferase from Campylobacter jejuni | | Descriptor: | SULFATE ION, Serine hydroxymethyltransferase | | Authors: | Anderson, S.M, Wawrzak, Z, Onopriyenko, O, Hasseman, J, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-05-14 | | Release date: | 2010-05-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of serine hydroxymethyltransferase from Campylobacter jejuni
TO BE PUBLISHED
|
|
6B75
 
 | | Crystal Structure of human NAMPT in complex with NVP-LOQ594 | | Descriptor: | 4-[(piperazin-1-yl)methyl]-N-{[4-({[(pyridin-3-yl)methyl]carbamoyl}amino)phenyl]methyl}benzamide, Nicotinamide phosphoribosyltransferase | | Authors: | Weihofen, W.A, Thigale, S. | | Deposit date: | 2017-10-03 | | Release date: | 2018-10-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Identification and structure based design of cellularly active cyclo-propyl carboxamide Nicotinamide phosphoribosyltransferase (NAMPT) inhibitors
To Be Published
|
|
6ATB
 
 | | Crystal Structure of human NAMPT in complex with NVP-LOD812 | | Descriptor: | DIMETHYL SULFOXIDE, GLYCEROL, N-{4-[(1,3-dioxo-1,3-dihydro-2H-isoindol-2-yl)methyl]phenyl}-N'-[(pyridin-3-yl)methyl]urea, ... | | Authors: | Weihofen, W.A, Thigale, S. | | Deposit date: | 2017-08-28 | | Release date: | 2018-09-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Identification and structure based design of cellularly active cyclo-propyl carboxamide Nicotinamide phosphoribosyltransferase (NAMPT) inhibitors
To Be Published
|
|
2H3B
 
 | | Crystal Structure of Mouse Nicotinamide Phosphoribosyltransferase/Visfatin/Pre-B Cell Colony Enhancing Factor 1 | | Descriptor: | Nicotinamide phosphoribosyltransferase, SULFATE ION | | Authors: | Wang, T, Zhang, X, Bheda, P, Revollo, J.R, Imai, S.I, Wolberger, C. | | Deposit date: | 2006-05-22 | | Release date: | 2006-06-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of Nampt/PBEF/visfatin, a mammalian NAD(+) biosynthetic enzyme.
Nat.Struct.Mol.Biol., 13, 2006
|
|
6AZJ
 
 | | Crystal Structure of human NAMPT in complex with NVP-LQN520 | | Descriptor: | (1S,2S)-N-{4-[(1,3-dioxo-1,3-dihydro-2H-isoindol-2-yl)methyl]phenyl}-2-(pyridin-3-yl)cyclopropane-1-carboxamide, Nicotinamide phosphoribosyltransferase | | Authors: | Weihofen, W.A, Thigale, S. | | Deposit date: | 2017-09-11 | | Release date: | 2018-09-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Identification and structure based design of cellularly active cyclo-propyl carboxamide Nicotinamide phosphoribosyltransferase (NAMPT) inhibitors
To Be Published
|
|
5G31
 
 | |
2H3D
 
 | | Crystal Structure of Mouse Nicotinamide Phosphoribosyltransferase/Visfatin/Pre-B Cell Colony Enhancing Factor in Complex with Nicotinamide Mononuleotide | | Descriptor: | BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, Nicotinamide phosphoribosyltransferase | | Authors: | Wang, T, Zhang, X, Bheda, P, Revollo, J.R, Imai, S.I, Wolberger, C. | | Deposit date: | 2006-05-22 | | Release date: | 2006-06-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of Nampt/PBEF/visfatin, a mammalian NAD(+) biosynthetic enzyme.
Nat.Struct.Mol.Biol., 13, 2006
|
|
3O2R
 
 | | Structural flexibility in region involved in dimer formation of nuclease domain of Ribonuclase III (rnc) from Campylobacter jejuni | | Descriptor: | CHLORIDE ION, Ribonuclease III | | Authors: | Minasov, G, Halavaty, A, Shuvalova, L, Dubrovska, I, Winsor, J, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-07-22 | | Release date: | 2010-08-04 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.251 Å) | | Cite: | Structural Flexibility in Region Involved in Dimer Formation of Nuclease Domain of Ribonuclase III (rnc) from Campylobacter jejuni.
TO BE PUBLISHED
|
|
3O6D
 
 | | Pyridoxal phosphate biosynthetic protein PdxJ from Campylobacter jejuni in complex with pyridoxine-5'-phosphate | | Descriptor: | PHOSPHATE ION, PYRIDOXINE-5'-PHOSPHATE, Pyridoxine 5'-phosphate synthase | | Authors: | Osipiuk, J, Zhou, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-07-28 | | Release date: | 2010-08-11 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | X-ray crystal structure of pyridoxal phosphate biosynthetic protein PdxJ from Campylobacter jejuni.
To be Published
|
|
1KRQ
 
 | | CRYSTAL STRUCTURE ANALYSIS OF CAMPYLOBACTER JEJUNI FERRITIN | | Descriptor: | ferritin | | Authors: | Hortolan, L, Saintout, N, Granier, G, Langlois d'Estaintot, B, Manigand, C, Mizunoe, Y, Wai, S.N, Gallois, B, Precigoux, G. | | Deposit date: | 2002-01-10 | | Release date: | 2002-02-06 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | STRUCTURE OF CAMPYLOBACTER JEJUNI FERRITIN AT 2.7 A RESOLUTION
To be Published
|
|
7PQF
 
 | | Crystal structure of Campylobacter jejuni DsbA2 | | Descriptor: | Thiol:disulfide interchange protein DsbA/DsbL | | Authors: | Wilk, P, Banas, A.M, Bocian-Ostrzycka, K.M, Jagusztyn-Krynicka, E.K. | | Deposit date: | 2021-09-17 | | Release date: | 2021-12-29 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Interplay between DsbA1, DsbA2 and C8J_1298 Periplasmic Oxidoreductases of Campylobacter jejuni and Their Impact on Bacterial Physiology and Pathogenesis.
Int J Mol Sci, 22, 2021
|
|
7PQ8
 
 | | Crystal structure of Campylobacter jejuni DsbA1 | | Descriptor: | TETRAETHYLENE GLYCOL, Thiol:disulfide interchange protein DsbA | | Authors: | Orlikowska, M, Bocian-Ostrzycka, K.M, Banas, A.M, Jagusztyn-Krynicka, E.K. | | Deposit date: | 2021-09-16 | | Release date: | 2021-12-29 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.329 Å) | | Cite: | Interplay between DsbA1, DsbA2 and C8J_1298 Periplasmic Oxidoreductases of Campylobacter jejuni and Their Impact on Bacterial Physiology and Pathogenesis.
Int J Mol Sci, 22, 2021
|
|
7PQ7
 
 | | Crystal structure of Campylobacter jejuni DsbA1 | | Descriptor: | TETRAETHYLENE GLYCOL, TRIETHYLENE GLYCOL, Thiol:disulfide interchange protein DsbA | | Authors: | Wilk, P, Orlikowska, M, Banas, A.M, Bocian-Ostrzycka, K.M, Jagusztyn-Krynicka, E.K. | | Deposit date: | 2021-09-16 | | Release date: | 2021-12-29 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Interplay between DsbA1, DsbA2 and C8J_1298 Periplasmic Oxidoreductases of Campylobacter jejuni and Their Impact on Bacterial Physiology and Pathogenesis.
Int J Mol Sci, 22, 2021
|
|
5YLL
 
 | | Structure of GH113 beta-1,4-mannanase complex with M6. | | Descriptor: | beta-1,4-mannanase, beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-beta-D-mannopyranose | | Authors: | Jiang, Z.Q, You, X, Yang, S.Q, Huang, P, Ma, J.W. | | Deposit date: | 2017-10-17 | | Release date: | 2018-06-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structural insights into the catalytic mechanism of a novel glycoside hydrolase family 113 beta-1,4-mannanase from Amphibacillus xylanus
J. Biol. Chem., 293, 2018
|
|
