9BRR
 
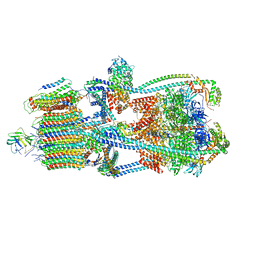 | | Intact V-ATPase State 3 in synaptophysin knock-out isolated synaptic vesicles | | Descriptor: | Renin receptor cytoplasmic fragment, Ribonuclease kappa, V-type proton ATPase 116 kDa subunit a 1, ... | | Authors: | Wang, C, Jiang, W, Yang, K, Wang, X, Guo, Q, Brunger, A.T. | | Deposit date: | 2024-05-11 | | Release date: | 2024-06-19 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structure and topography of the synaptic V-ATPase-synaptophysin complex.
Nature, 631, 2024
|
|
9BRQ
 
 | | Intact V-ATPase State 3 and synaptophysin complex in mouse brain isolated synaptic vesicles | | Descriptor: | Renin receptor cytoplasmic fragment, Ribonuclease kappa, Synaptophysin, ... | | Authors: | Wang, C, Jiang, W, Yang, K, Wang, X, Guo, Q, Brunger, A.T. | | Deposit date: | 2024-05-11 | | Release date: | 2024-06-19 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structure and topography of the synaptic V-ATPase-synaptophysin complex.
Nature, 631, 2024
|
|
9BRD
 
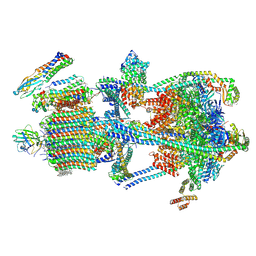 | | Synaptic Vesicle V-ATPase with synaptophysin and SidK, State 3 | | Descriptor: | (7R)-4,7-DIHYDROXY-N,N,N-TRIMETHYL-10-OXO-3,5,9-TRIOXA-4-PHOSPHAHEPTACOSAN-1-AMINIUM 4-OXIDE, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Coupland, C.E, Rubinstein, J.L. | | Deposit date: | 2024-05-11 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | High-resolution electron cryomicroscopy of V-ATPase in native synaptic vesicles.
Science, 385, 2024
|
|
9BRC
 
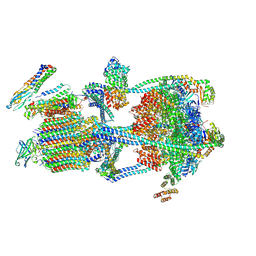 | |
9BRB
 
 | |
9BRA
 
 | | Intact V-ATPase State 2 and synaptophysin complex in mouse brain isolated synaptic vesicles | | Descriptor: | Renin receptor cytoplasmic fragment, Ribonuclease kappa, Synaptophysin, ... | | Authors: | Wang, C, Jiang, W, Yang, K, Wang, X, Guo, Q, Brunger, A.T. | | Deposit date: | 2024-05-11 | | Release date: | 2024-06-19 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structure and topography of the synaptic V-ATPase-synaptophysin complex.
Nature, 631, 2024
|
|
9BR7
 
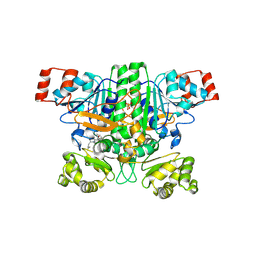 | | Crystal structure of human succinyl-CoA:glutarate-CoA transferase (SUGCT) in complex with Losartan carboxylic acid | | Descriptor: | AMMONIUM ION, SULFATE ION, Succinate--hydroxymethylglutarate CoA-transferase, ... | | Authors: | Wu, R, Lazarus, M.B. | | Deposit date: | 2024-05-10 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Characterization, Structure, and Inhibition of the Human Succinyl-CoA:glutarate-CoA Transferase, a Putative Genetic Modifier of Glutaric Aciduria Type 1.
Acs Chem.Biol., 19, 2024
|
|
9BR6
 
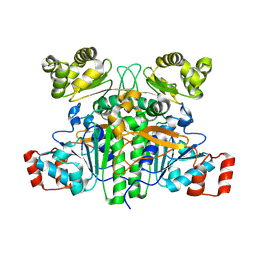 | |
9BQV
 
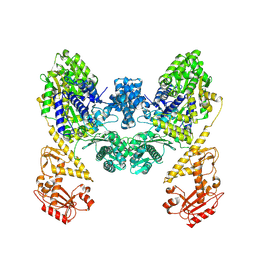 | | DdmD dimer apoprotein | | Descriptor: | Helicase/UvrB N-terminal domain-containing protein | | Authors: | Bravo, J.P.K, Taylor, D.W. | | Deposit date: | 2024-05-10 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Plasmid targeting and destruction by the DdmDE bacterial defence system.
Nature, 630, 2024
|
|
9BQJ
 
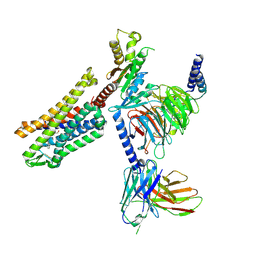 | | RO76 bound muOR-Gi1-scFv16 complex structure | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Wang, H, Majumdar, S, Kobilka, B.K. | | Deposit date: | 2024-05-10 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Signaling Modulation Mediated by Ligand Water Interactions with the Sodium Site at mu OR.
Acs Cent.Sci., 10, 2024
|
|
9BQE
 
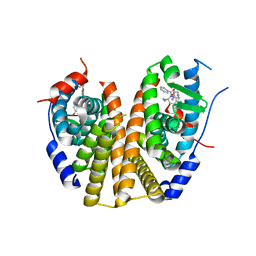 | | Estrogen Receptor Alpha Ligand Binding Domain Y537S Mutant in Complex with (6'-hydroxy-1'-(4-(2-(1-propylpyrrolidin-3-yl)ethoxy)phenyl)-1',4'-dihydro-2'H-spiro[cyclopropane-1,3'-isoquinolin]-2'-yl)(phenyl)methanone | | Descriptor: | Estrogen receptor, [(1'R)-6'-hydroxy-1'-(4-{[(3R)-1-propylpyrrolidin-3-yl]methoxy}phenyl)-1',4'-dihydro-2'H-spiro[cyclopropane-1,3'-isoquinolin]-2'-yl](phenyl)methanone | | Authors: | Young, K.Y, Fanning, S.W. | | Deposit date: | 2024-05-09 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Estrogen Receptor Alpha Ligand Binding Domain Y537S Mutant in Complex with (6'-hydroxy-1'-(4-(2-(1-propylpyrrolidin-3-yl)ethoxy)phenyl)-1',4'-dihydro-2'H-spiro[cyclopropane-1,3'-isoquinolin]-2'-yl)(phenyl)methanone
To Be Published
|
|
9BQ4
 
 | |
9BQ2
 
 | |
9BQ0
 
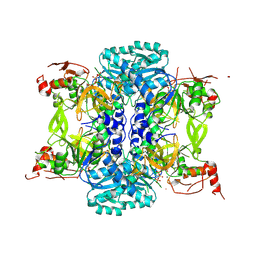 | |
9BPF
 
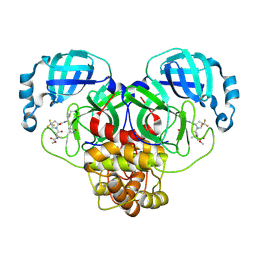 | | Crystal structure of main protease of SARS-CoV-2 complexed with inhibitor | | Descriptor: | 3C-like proteinase nsp5, N-(methoxycarbonyl)-3-methyl-L-valyl-(4R)-N-{(1Z,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-4-(trifluoromethyl)-L-prolinamide | | Authors: | Chen, P, Arutyunova, E, Lemieux, M.J. | | Deposit date: | 2024-05-07 | | Release date: | 2024-08-07 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Structural Comparison of Oral SARS-CoV-2 Drug Candidate Ibuzatrelvir Complexed with the Main Protease (M pro ) of SARS-CoV-2 and MERS-CoV.
Jacs Au, 4, 2024
|
|
9BPE
 
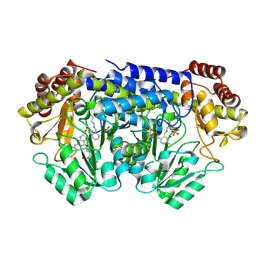 | | Joint X-ray/neutron structure of Thermus thermophilus serine hydroxymethyltransferase (TthSHMT) in internal aldimine state and folinic acid bound | | Descriptor: | ACETATE ION, N-[4-({[(6S)-2-amino-5-formyl-4-oxo-3,4,5,6,7,8-hexahydropteridin-6-yl]methyl}amino)benzoyl]-L-glutamic acid, SULFATE ION, ... | | Authors: | Drago, V.N, Kovalevsky, A. | | Deposit date: | 2024-05-07 | | Release date: | 2024-08-28 | | Method: | NEUTRON DIFFRACTION (2.3 Å), X-RAY DIFFRACTION | | Cite: | Universality of critical active site glutamate as an acid-base catalyst in serine hydroxymethyltransferase function.
Chem Sci, 15, 2024
|
|
9BPA
 
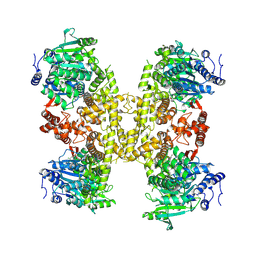 | | Human DNA polymerase theta helicase domain in complex with inhibitor AB25583, tetramer form | | Descriptor: | (4P)-N-{5-[(4-chlorophenyl)methoxy]-1,3,4-thiadiazol-2-yl}-4-(2-methoxyphenyl)pyridine-3-carboxamide, DNA polymerase theta | | Authors: | Ito, F, Li, Z, Chen, X.S. | | Deposit date: | 2024-05-07 | | Release date: | 2024-08-28 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Structural basis for a Pol theta helicase small-molecule inhibitor revealed by cryo-EM.
Nat Commun, 15, 2024
|
|
9BP9
 
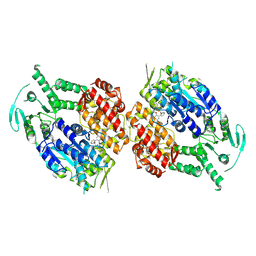 | | Human DNA polymerase theta helicase domain in complex with inhibitor AB25583, dimer form | | Descriptor: | (4P)-N-{5-[(4-chlorophenyl)methoxy]-1,3,4-thiadiazol-2-yl}-4-(2-methoxyphenyl)pyridine-3-carboxamide, DNA polymerase theta | | Authors: | Ito, F, Li, Z, Chen, X.S. | | Deposit date: | 2024-05-07 | | Release date: | 2024-08-28 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Structural basis for a Pol theta helicase small-molecule inhibitor revealed by cryo-EM.
Nat Commun, 15, 2024
|
|
9BP6
 
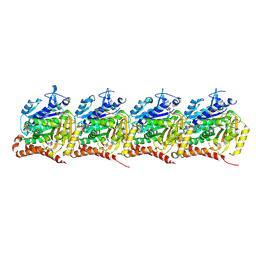 | |
9BOX
 
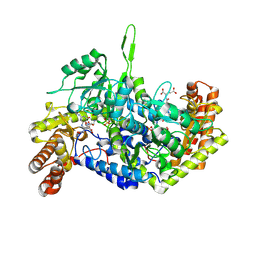 | | Room-temperature X-ray structure of human mitochondrial serine hydroxymethyltransferase (hSHMT2) with PLP-glycine external aldimine and 5-formyltetrahydrofolate (folinic acid) | | Descriptor: | (E)-N-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)glycine, N-[4-({[(6S)-2-amino-5-formyl-4-oxo-3,4,5,6,7,8-hexahydropteridin-6-yl]methyl}amino)benzoyl]-L-glutamic acid, Serine hydroxymethyltransferase, ... | | Authors: | Drago, V.N, Kovalevsky, A. | | Deposit date: | 2024-05-06 | | Release date: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Universality of critical active site glutamate as an acid-base catalyst in serine hydroxymethyltransferase function.
Chem Sci, 15, 2024
|
|
9BOW
 
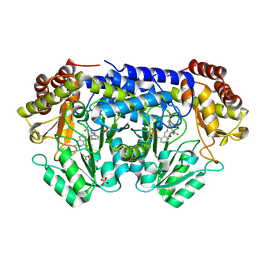 | | X-ray structure of Thermus thermophilus serine hydroxymethyltransferase with PLP-L-Ser external aldimine and 5-formyltetrahydrofolate (folinic acid) | | Descriptor: | (E)-N-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)-L-serine, N-[4-({[(6S)-2-amino-5-formyl-4-oxo-3,4,5,6,7,8-hexahydropteridin-6-yl]methyl}amino)benzoyl]-L-glutamic acid, SERINE, ... | | Authors: | Drago, V.N, Kovalevsky, A. | | Deposit date: | 2024-05-06 | | Release date: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Universality of critical active site glutamate as an acid-base catalyst in serine hydroxymethyltransferase function.
Chem Sci, 15, 2024
|
|
9BOR
 
 | | IkappaBzeta ankyrin repeat domain:NF-kappaB p50 homodimer complex at 2.0 Angstrom resolution | | Descriptor: | (2R,3S)-heptane-1,2,3-triol, NF-kappa-B inhibitor zeta, Nuclear factor NF-kappa-B p50 subunit | | Authors: | Rogers, W.E, Zhu, N, Huxford, T. | | Deposit date: | 2024-05-05 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | Structural and biochemical analyses of the nuclear I kappa B zeta protein in complex with the NF-kappa B p50 homodimer.
Genes Dev., 38, 2024
|
|
9BOQ
 
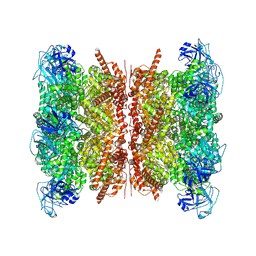 | | Human p97/VCP structure with a triazole inhibitor (NSC799462/dodecamer) | | Descriptor: | 3-(2,6-difluoro-4-{[(4P)-5-{[(2S)-hexan-2-yl]sulfanyl}-4-(pyridin-3-yl)-4H-1,2,4-triazol-3-yl]methoxy}phenyl)prop-2-yn-1-yl (1-methylpiperidin-4-yl)carbamate, ADENOSINE-5'-DIPHOSPHATE, Transitional endoplasmic reticulum ATPase | | Authors: | Nandi, P, DeVore, K, Chiu, P.-L. | | Deposit date: | 2024-05-05 | | Release date: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | Mechanism of allosteric inhibition of human p97/VCP ATPase and its disease mutant by triazole inhibitors.
Commun Chem, 7, 2024
|
|
9BOO
 
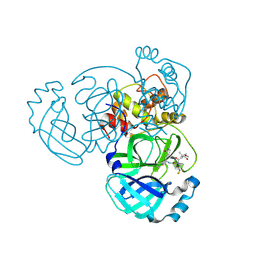 | | Crystal structure of MERS-CoV Nsp5 in complex with PF-07817883 | | Descriptor: | 3C-like proteinase nsp5, N-(methoxycarbonyl)-3-methyl-L-valyl-(4R)-N-{(1Z,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-4-(trifluoromethyl)-L-prolinamide | | Authors: | Chen, P, Arutyunova, E, Lemieux, M.J. | | Deposit date: | 2024-05-05 | | Release date: | 2024-08-07 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A Structural Comparison of Oral SARS-CoV-2 Drug Candidate Ibuzatrelvir Complexed with the Main Protease (M pro ) of SARS-CoV-2 and MERS-CoV.
Jacs Au, 4, 2024
|
|
9BON
 
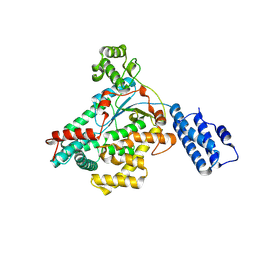 | | Crystal structure of glucosyltransferase (GTD) domain of TpeL | | Descriptor: | TpeL | | Authors: | Gill, S, Sugiman-Marangos, S.N, Beilhartz, G.L, Mei, E, Taipale, M, Melnyk, R.A. | | Deposit date: | 2024-05-05 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.222 Å) | | Cite: | A diphtheria toxin-like intracellular delivery platform that evades pre-existing antidrug antibodies.
Embo Mol Med, 2024
|
|
